

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.3 + phase: 0
(368 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
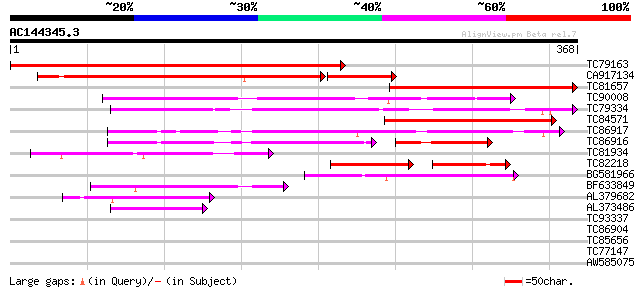
TC79163 weakly similar to PIR|E96724|E96724 hypothetical protein... 460 e-130
CA917134 weakly similar to PIR|E96724|E96 hypothetical protein F... 248 1e-80
TC81657 weakly similar to PIR|T51957|T51957 metalloproteinase (E... 243 9e-65
TC90008 weakly similar to GP|4249376|gb|AAD14473.1| Strong simil... 200 8e-52
TC79334 MtN9 158 4e-39
TC84571 weakly similar to SP|P29136|MEP1_SOYBN Metalloendoprotei... 150 1e-36
TC86917 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago trun... 145 2e-35
TC86916 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago trun... 96 1e-29
TC81934 weakly similar to PIR|E71433|E71433 probable metalloprot... 103 8e-23
TC82218 similar to GP|7159629|emb|CAB76364.1 matrix metalloprote... 62 3e-19
BG581966 similar to GP|3776080|emb MtN9 {Medicago truncatula}, p... 89 4e-18
BF633849 weakly similar to PIR|E71433|E714 probable metalloprote... 83 2e-16
AL379682 weakly similar to SP|P29136|MEP1_ Metalloendoproteinase... 64 9e-11
AL373486 weakly similar to GP|16901508|gb| matrix metalloprotein... 55 3e-08
TC93337 weakly similar to GP|21240660|gb|AAM44371.1 hypothetical... 31 0.67
TC86904 similar to GP|11908126|gb|AAG41492.1 unknown protein {Ar... 29 3.3
TC85656 homologue to PIR|S57786|S57786 phosphogluconate dehydrog... 28 4.3
TC77147 similar to GP|21703124|gb|AAM74503.1 AT3g13460/MRP15_10 ... 28 5.7
AW585075 28 7.4
>TC79163 weakly similar to PIR|E96724|E96724 hypothetical protein F20P5.11
[imported] - Arabidopsis thaliana, partial (32%)
Length = 762
Score = 460 bits (1183), Expect = e-130
Identities = 218/218 (100%), Positives = 218/218 (100%)
Frame = +2
Query: 1 MITQNQRYYYHIFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSR 60
MITQNQRYYYHIFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSR
Sbjct: 107 MITQNQRYYYHIFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSR 286
Query: 61 GKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATM 120
GKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATM
Sbjct: 287 GKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATM 466
Query: 121 NYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDP 180
NYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDP
Sbjct: 467 NYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDP 646
Query: 181 SENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI 218
SENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI
Sbjct: 647 SENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI 760
>CA917134 weakly similar to PIR|E96724|E96 hypothetical protein F20P5.11
[imported] - Arabidopsis thaliana, partial (26%)
Length = 723
Score = 248 bits (633), Expect(2) = 1e-80
Identities = 115/194 (59%), Positives = 144/194 (73%), Gaps = 7/194 (3%)
Frame = +1
Query: 19 SITLTSVSARFFPDPSSMPAW-NSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLKNYFNHF 77
S+TL VSAR FPD +P+W P G WD ++NFTGC +G+ Y+GLS LKNYF HF
Sbjct: 1 SLTLLIVSARLFPD---VPSWIPPGTLPVGPWDAFRNFTGCRQGENYNGLSNLKNYFQHF 171
Query: 78 GYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADIINGT 137
GYIP P SNF+DDFD+ L+ A++TYQKNFNLN+TGELDD T+ ++ PRCGVADIINGT
Sbjct: 172 GYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGT 351
Query: 138 TSMNSGKFNSSSTN------FHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDATKQV 191
T+MN+GK +++N FHTV+H++ FPGQPRWPEG Q LTYAF P L + K V
Sbjct: 352 TTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSV 531
Query: 192 FANAFNQWSKVTTI 205
FA AF +WS+VTT+
Sbjct: 532 FATAFARWSEVTTL 573
Score = 70.1 bits (170), Expect(2) = 1e-80
Identities = 32/45 (71%), Positives = 36/45 (79%)
Frame = +3
Query: 207 FTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRL 251
FTE T YS +DIKIGF GDHGDGE FDG LGTLAHAFS +G++
Sbjct: 576 FTETTLYSGADIKIGFLHGDHGDGEPFDGSLGTLAHAFSQGNGKV 710
>TC81657 weakly similar to PIR|T51957|T51957 metalloproteinase (EC 3.4.24.-)
[imported] - Arabidopsis thaliana, partial (24%)
Length = 537
Score = 243 bits (620), Expect = 9e-65
Identities = 122/122 (100%), Positives = 122/122 (100%)
Frame = +2
Query: 247 TDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISS 306
TDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISS
Sbjct: 2 TDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISS 181
Query: 307 RTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLFLSLVFLGFGFL 366
RTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLFLSLVFLGFGFL
Sbjct: 182 RTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLFLSLVFLGFGFL 361
Query: 367 SL 368
SL
Sbjct: 362 SL 367
>TC90008 weakly similar to GP|4249376|gb|AAD14473.1| Strong similarity to
GP|2829864 F3I6.6 zinc metalloproteinase homolog from
Arabidopsis thaliana BAC, partial (21%)
Length = 1173
Score = 200 bits (508), Expect = 8e-52
Identities = 121/275 (44%), Positives = 162/275 (58%), Gaps = 7/275 (2%)
Frame = +2
Query: 61 GKTYDGLSKLKNYFNHFGYIPNGP-PSNFTDDFDEALESAVRTYQKNFNLNITGELDDAT 119
G G+S LK Y FGY+ + SNFTD F E L+SA+ +Q NFNLN TG+LD
Sbjct: 254 GDIASGISDLKEYLQFFGYLNSSTLHSNFTDAFTENLQSAIIEFQTNFNLNTTGQLDQDI 433
Query: 120 MNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPE-GTQTLTYAF 178
I KPRCGV DIINGTT+M + N + P +P W ++L YAF
Sbjct: 434 YKIISKPRCGVPDIINGTTTMKNNFINKT------------MPFKPWWRNVENRSLAYAF 577
Query: 179 DPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLG 238
P N+ D K +F +AFN+WS T + F E S++ SDI+I F + DG G
Sbjct: 578 HPENNVTDNVKSLFQDAFNRWSNATELNFIETMSFNDSDIRIAFLT--------LDGKGG 733
Query: 239 TLAHAF---SPTDGRLHLDKAEDWVV-NGDVTESSLSNAVDLESVVVHEIGHLLGLGHSS 294
T+ ++ S G ++LD E WV+ + +V E + VDLESVV+H++GHLLGLGHSS
Sbjct: 734 TVGGSYINSSVNVGSVYLDADEQWVLPSENVVE---EDDVDLESVVMHQVGHLLGLGHSS 904
Query: 295 IEEAIMYPTISSRTKKVELES-DDIEGIQMLYGSN 328
+EEAIMYP I + KK+EL + DD++ IQ +YG N
Sbjct: 905 VEEAIMYP-IVLQEKKIELVNVDDLQRIQEIYGVN 1006
>TC79334 MtN9
Length = 1241
Score = 158 bits (399), Expect = 4e-39
Identities = 111/311 (35%), Positives = 155/311 (49%), Gaps = 8/311 (2%)
Frame = +1
Query: 66 GLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVK 125
GLSK+K + HF Y+ F D D SA++ YQ+ FNL +TG LD T+ I+
Sbjct: 112 GLSKIKQHLYHFKYLQGLYLVGFDDYLDNKTISAIKAYQQFFNLQVTGHLDTETLQQIML 291
Query: 126 PRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLD 185
PRCGV D IN + + G ++ G +P+GT+ LTY F P +
Sbjct: 292 PRCGVPD-INPDINPDFG--------------FARAQGNKWFPKGTKELTYGFLPESKIS 426
Query: 186 DATKQVFANAFNQWSKVT-TITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAF 244
VF NAF +WS+ T + F+EATSY +DIKIGFY+ + E D V+
Sbjct: 427 IDKVNVFRNAFTRWSQTTRVLKFSEATSYDDADIKIGFYNISYNSKEVIDVVVSDF--FI 600
Query: 245 SPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTI 304
+ + L+ ++ W DLE+V +H+IGHLLGL HSS E+IMYPTI
Sbjct: 601 NLRSFTIRLEASKVW---------------DLETVAMHQIGHLLGLDHSSDVESIMYPTI 735
Query: 305 -SSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDS----SFGGR--HGVSLFLS 357
KKV++ D + IQ LY T T++DRD + G S L+
Sbjct: 736 VPLHQKKVQITVSDNQAIQQLY---------TKQTNQDRDELGFFDYSGDFFESSSGLLN 888
Query: 358 LVFLGFGFLSL 368
+ LGF F++L
Sbjct: 889 SLSLGFAFVAL 921
>TC84571 weakly similar to SP|P29136|MEP1_SOYBN Metalloendoproteinase 1
precursor (EC 3.4.24.-) (SMEP1). [Soybean] {Glycine
max}, partial (28%)
Length = 506
Score = 150 bits (378), Expect = 1e-36
Identities = 75/112 (66%), Positives = 88/112 (77%)
Frame = +2
Query: 244 FSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPT 303
FSP +GR HLD AEDWVV+GDV++SSL AVDLESV VHEIGHLLGLGHSS EEAIM+PT
Sbjct: 2 FSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPT 181
Query: 304 ISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLF 355
ISSR KKV L DD+ GIQ LYG+NP+F G+T +S +R+ GG SL+
Sbjct: 182 ISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLW 337
>TC86917 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago truncatula},
partial (59%)
Length = 1294
Score = 145 bits (367), Expect = 2e-35
Identities = 106/306 (34%), Positives = 149/306 (48%), Gaps = 9/306 (2%)
Frame = +1
Query: 64 YDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYI 123
Y G+ ++K Y + FGY+ P N T D + L A++TYQ+ FN I + + +I
Sbjct: 337 YKGVDQIKQYLSDFGYLEQSGPFNNTLDQETVL--ALKTYQRYFN--IQQDTLSEILQHI 504
Query: 124 VKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSEN 183
PRCGV D I K+N ++ SF G +P+GT+ LTY FDP
Sbjct: 505 ALPRCGVPDRIL--------KYNLTND-------ISFPKGNQWFPKGTKNLTYGFDPRNK 639
Query: 184 LDDATKQVFANAFNQWSKVTTI-TFTEATSYSSSDIKIGFYS--GDHGDGEAFDGVLGTL 240
+ VF A QWS T + FTE SY ++IKIGFY+ D G + G +
Sbjct: 640 IPLDMTNVFRTALTQWSNTTRVLNFTETKSYDDANIKIGFYNITDDDGINDVAVGFTFIV 819
Query: 241 AHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIM 300
+ + G + LD + W + + DLE+ +H+IGHLLGL HSS ++IM
Sbjct: 820 LDSTNVKSGFITLDATKYWALPTE------HRGFDLETAAMHQIGHLLGLEHSSDNKSIM 981
Query: 301 YPTI-SSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSS-----FGGRHGVSL 354
YPTI S K V++ D IQ LY S +T A + DSS FG + +
Sbjct: 982 YPTILPSHQKNVQITDSDNLAIQKLYSS------STKANANSDDSSGCFKLFGSSSSLLI 1143
Query: 355 FLSLVF 360
LS+VF
Sbjct: 1144SLSIVF 1161
>TC86916 similar to GP|3776080|emb|CAA77093.1 MtN9 {Medicago truncatula},
partial (39%)
Length = 954
Score = 96.3 bits (238), Expect(2) = 1e-29
Identities = 62/176 (35%), Positives = 88/176 (49%), Gaps = 1/176 (0%)
Frame = +2
Query: 64 YDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYI 123
Y GL ++K Y +FGY+ P N T D + L A++TYQ+ FN+ + + +I
Sbjct: 260 YKGLDQIKQYLQNFGYLEQSGPFNNTLDQETVL--ALKTYQRYFNIYAGQDSLRKILQHI 433
Query: 124 VKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSEN 183
PRCGV D+ F STN S+ G +P+GT+ LTY F P
Sbjct: 434 ALPRCGVPDM----------NFTYDSTN-----DISYPKGNQWFPKGTKNLTYGFAPKNE 568
Query: 184 LDDATKQVFANAFNQWSKVT-TITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLG 238
+ VF A +WS+ T + FTE TSY +DIKI F + + DG +D V+G
Sbjct: 569 IPLNVTNVFRKALTRWSQTTRVLNFTETTSYDDADIKIVFNNMTYDDG-IYDVVVG 733
Score = 51.2 bits (121), Expect(2) = 1e-29
Identities = 29/64 (45%), Positives = 41/64 (63%), Gaps = 1/64 (1%)
Frame = +1
Query: 251 LHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTI-SSRTK 309
+ LD + WV + E +DLE+ +H+IGHLLGL HSS ++IMYPTI S+ K
Sbjct: 781 ISLDITKHWVFPTEDGE------LDLETAAMHQIGHLLGLEHSSDSKSIMYPTILPSQQK 942
Query: 310 KVEL 313
KV++
Sbjct: 943 KVQI 954
>TC81934 weakly similar to PIR|E71433|E71433 probable metalloproteinase (EC
3.4.24.-) - Arabidopsis thaliana, partial (24%)
Length = 619
Score = 103 bits (258), Expect = 8e-23
Identities = 58/164 (35%), Positives = 83/164 (50%), Gaps = 6/164 (3%)
Frame = +1
Query: 14 YITLISITLTSVSARFFPD---PSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKL 70
YI + +T FP P S+ ++ W + F G G+++L
Sbjct: 52 YINFLFFFMTLFLRPCFPARIIPESVTVITTTQKHNATWHDFSRFLHAEPGSHVSGMAEL 231
Query: 71 KNYFNHFGYIPNGPP---SNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPR 127
K YFN FGY+ PP +NFTD FD + +SAV YQKN L ITG+LD T++ IV PR
Sbjct: 232 KKYFNRFGYL--SPPLTTANFTDTFDSSFQSAVVLYQKNLGLPITGKLDSNTISTIVSPR 405
Query: 128 CGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGT 171
CGV+D +++ HT H+++F G+PRW G+
Sbjct: 406 CGVSD-------------TAATHRIHTTQHFAYFNGKPRWLRGS 498
>TC82218 similar to GP|7159629|emb|CAB76364.1 matrix metalloproteinase
{Cucumis sativus}, partial (19%)
Length = 688
Score = 62.0 bits (149), Expect(2) = 3e-19
Identities = 29/54 (53%), Positives = 33/54 (60%)
Frame = +3
Query: 209 EATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVN 262
E S+DIKIGF+ G+HGD FDG LAH F P DGRLH D E+W N
Sbjct: 3 EVGDEGSADIKIGFHRGNHGDVYPFDGPGNVLAHTFPPEDGRLHFDGDENWTNN 164
Score = 50.4 bits (119), Expect(2) = 3e-19
Identities = 24/51 (47%), Positives = 34/51 (66%)
Frame = +1
Query: 275 DLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLY 325
DLE+V +HE+GHLLGL HS+ + + MYP + + L DD++GI LY
Sbjct: 262 DLETVALHEMGHLLGLAHSTDQNSAMYPYWAGVRR--NLNQDDVDGITALY 408
>BG581966 similar to GP|3776080|emb MtN9 {Medicago truncatula}, partial (53%)
Length = 683
Score = 88.6 bits (218), Expect = 4e-18
Identities = 60/150 (40%), Positives = 84/150 (56%), Gaps = 11/150 (7%)
Frame = +1
Query: 192 FANAFNQWSKVTTI-TFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHA----FSP 246
F NAF +WS+ T + F+E TSY +DIKIGFY + D D V+G + +
Sbjct: 19 FRNAFTRWSQTTRVLNFSETTSYDDADIKIGFYHIYNND-IVDDVVVGDSFISRNLDSNV 195
Query: 247 TDGRLHLDKAEDWVVNGDVTESSLS-NAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTIS 305
G + L++++ WV DLE+VV+H+IGHLLGL HSS +E+IMYP I
Sbjct: 196 KSGMIRLERSKFWVSPTTTYFKKWELQEFDLETVVMHQIGHLLGLDHSSDKESIMYPLID 375
Query: 306 S-RTKKVELESDDIEGIQMLY----GSNPN 330
+ KKV++ D + IQ LY +NPN
Sbjct: 376 PLQEKKVQITDSDNQAIQQLYTNTAKANPN 465
>BF633849 weakly similar to PIR|E71433|E714 probable metalloproteinase (EC
3.4.24.-) - Arabidopsis thaliana, partial (5%)
Length = 544
Score = 82.8 bits (203), Expect = 2e-16
Identities = 43/133 (32%), Positives = 68/133 (50%), Gaps = 4/133 (3%)
Frame = +2
Query: 53 KNFTGCSRGKTYDGLSKLKNYFNHFGYI----PNGPPSNFTDDFDEALESAVRTYQKNFN 108
+N G +G+T G+ +LKNY FGY+ P + D FDE LE A++ +Q +
Sbjct: 149 QNLKGVQKGQTQKGIGELKNYLKRFGYLNVQHDATPSTTLNDHFDENLEFALKDFQTYHH 328
Query: 109 LNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWP 168
L++TG +D T+ + PRCGV D+ + N + +SS Y+FF P+W
Sbjct: 329 LHVTGRVDTTTIKILSLPRCGVPDLPKHSHKQNGLQMSSS---------YAFFQDSPKWS 481
Query: 169 EGTQTLTYAFDPS 181
+ + L Y + S
Sbjct: 482 DTKRNLKYMYKSS 520
>AL379682 weakly similar to SP|P29136|MEP1_ Metalloendoproteinase 1 precursor
(EC 3.4.24.-) (SMEP1). [Soybean] {Glycine max}, partial
(15%)
Length = 386
Score = 63.9 bits (154), Expect = 9e-11
Identities = 40/102 (39%), Positives = 53/102 (51%), Gaps = 3/102 (2%)
Frame = +3
Query: 35 SMPAWNSSNAPPGAWDGYKNFTGCSRGKTYD---GLSKLKNYFNHFGYIPNGPPSNFTDD 91
S P+ +S + P GY S GK + GLSK+K + HF Y+ F D
Sbjct: 6 SYPSSSSLSIQP--LSGYIPQLSPSLGKQTEEIQGLSKIKQHLYHFKYLQGLYLVGFDDY 179
Query: 92 FDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADI 133
D SA++ YQ+ FNL +TG LD T+ I+ PRCGV DI
Sbjct: 180 LDNKTISAIKAYQQFFNLQVTGHLDTETLQQIMLPRCGVPDI 305
>AL373486 weakly similar to GP|16901508|gb| matrix metalloproteinase MMP2
{Glycine max}, partial (7%)
Length = 409
Score = 55.5 bits (132), Expect = 3e-08
Identities = 27/63 (42%), Positives = 36/63 (56%)
Frame = +2
Query: 66 GLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVK 125
GLSK+K + HF Y+ F D D SA++ YQ+ FNL +TG LD T+ I+
Sbjct: 119 GLSKIKQHLYHFKYLQGLYLVGFDDYLDNKTISAIKGYQQFFNLQVTGHLDTETLQQIML 298
Query: 126 PRC 128
PRC
Sbjct: 299 PRC 307
>TC93337 weakly similar to GP|21240660|gb|AAM44371.1 hypothetical protein
{Dictyostelium discoideum}, partial (7%)
Length = 743
Score = 31.2 bits (69), Expect = 0.67
Identities = 38/174 (21%), Positives = 73/174 (41%), Gaps = 6/174 (3%)
Frame = +1
Query: 169 EGTQTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI--KIGFYSGD 226
EG Q L + + E ++ K A++ TT T TE S +SD+ ++G +
Sbjct: 208 EGNQGLVESNNIHEAREEQYKGDDASSAVTHDTRTTSTETETVSMENSDVNAEMGITKPE 387
Query: 227 HGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAV----DLESVVVH 282
+ DG+ FS T+ +L + + + + +SLSN V + +++
Sbjct: 388 NKPNYTEDGIRSQPGSNFSITEVKLVVGAYSNSSSSNETGNNSLSNPVAGSHQNNTALIY 567
Query: 283 EIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTT 336
+ H +S I+ P + + S+ + + M+ G + N TG +T
Sbjct: 568 SVRH---SEETSSNLTIVVPAAKNNMSGTDASSEHHKMV-MVSGLDKNDTGNST 717
>TC86904 similar to GP|11908126|gb|AAG41492.1 unknown protein {Arabidopsis
thaliana}, partial (42%)
Length = 2062
Score = 28.9 bits (63), Expect = 3.3
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = -2
Query: 8 YYYHIFYITLISITLTSVSARFF 30
Y+YH F+I++I L+ SAR F
Sbjct: 1893 YFYHRFFISIIFFLLSKASARIF 1825
>TC85656 homologue to PIR|S57786|S57786 phosphogluconate dehydrogenase
(decarboxylating) (EC 1.1.1.44) - alfalfa, complete
Length = 1811
Score = 28.5 bits (62), Expect = 4.3
Identities = 18/66 (27%), Positives = 30/66 (45%)
Frame = +3
Query: 172 QTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGE 231
Q + A+D +++ T + +AF +W+K ++F + IK D GDG
Sbjct: 711 QLIAEAYDVLKSVGKLTNEELQSAFTEWNKGELLSFLIEITADIFGIK-----DDKGDGY 875
Query: 232 AFDGVL 237
D VL
Sbjct: 876 LVDKVL 893
>TC77147 similar to GP|21703124|gb|AAM74503.1 AT3g13460/MRP15_10
{Arabidopsis thaliana}, partial (32%)
Length = 2310
Score = 28.1 bits (61), Expect = 5.7
Identities = 19/63 (30%), Positives = 24/63 (37%), Gaps = 8/63 (12%)
Frame = +2
Query: 29 FFPDPSSMPAWNSSNA------PPGAWDGYKNFTGCSRGKTYDGLSKLKNYFNH--FGYI 80
F P+PS +P S G W GY N+ G T +Y H +GY
Sbjct: 242 FNPNPSYVPNGYPSTYYYGGYDGQGDWSGYSNYVNLDGGMTQGVYGDNCSYMYHQGYGYT 421
Query: 81 PNG 83
P G
Sbjct: 422 PYG 430
>AW585075
Length = 406
Score = 27.7 bits (60), Expect = 7.4
Identities = 16/39 (41%), Positives = 20/39 (51%)
Frame = +3
Query: 166 RWPEGTQTLTYAFDPSENLDDATKQVFANAFNQWSKVTT 204
R+P G + T AF+PS N D + VF N K TT
Sbjct: 141 RYPSGNKVTTRAFNPSGNAD--AQPVFQVTLNVAPKQTT 251
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,606,316
Number of Sequences: 36976
Number of extensions: 168595
Number of successful extensions: 913
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 891
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 894
length of query: 368
length of database: 9,014,727
effective HSP length: 97
effective length of query: 271
effective length of database: 5,428,055
effective search space: 1471002905
effective search space used: 1471002905
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144345.3


