

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.13 + phase: 0
(471 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
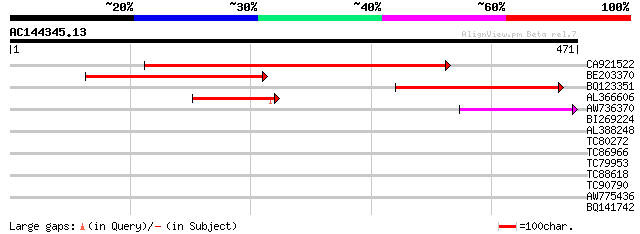
Score E
Sequences producing significant alignments: (bits) Value
CA921522 289 2e-78
BE203370 274 5e-74
BQ123351 105 4e-23
AL366606 81 8e-16
AW736370 51 1e-06
BI269224 weakly similar to GP|11862972|db hypothetical protein {... 31 0.90
AL388248 weakly similar to GP|6062758|gb|A NADH dehydrogenase su... 30 2.6
TC80272 similar to GP|15620810|dbj|BAB67768. RPS6-like protein {... 29 4.4
TC86966 similar to GP|6850934|emb|CAB71030.1 lectin-like protein... 29 4.4
TC79953 similar to GP|20279471|gb|AAM18751.1 unknown protein {Or... 29 4.4
TC88618 weakly similar to PIR|T48397|T48397 S-receptor kinase-li... 28 5.8
TC90790 similar to GP|18086412|gb|AAL57663.1 At2g17250/T23A1.11 ... 28 5.8
AW775436 28 7.6
BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvo... 28 7.6
>CA921522
Length = 767
Score = 289 bits (739), Expect = 2e-78
Identities = 138/254 (54%), Positives = 178/254 (69%)
Frame = +3
Query: 113 KTSLIKEKLTLKGNLPSLPTKFLYQQAFNFSKTNNVEAFYSILALLIYGLVLFPNIDNYV 172
+T +K L +G T FLY++ F K AF SILALL+YGLVLFP++DN+V
Sbjct: 3 ETDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFV 182
Query: 173 DIHAIQIFLTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHLPQTHSFQAN 232
DI AIQIFL++NPVPTLL D Y SIH RTQ GRG IL CA LLY+W TSHLP+T F N
Sbjct: 183 DIKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPRFTTN 362
Query: 233 PENLSWPKRIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPILAKRQF 292
PENL W KR+MSLTP+++ WY + II SCG+++NVPLLG +GGISYNP LA+ QF
Sbjct: 363 PENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARHQF 542
Query: 293 GCPMEAKPDNIYLQGEFYFNHEDPSNKRGRFVQAWHAIRTLNRSQLARRSDSLQGSYTQW 352
G PME KP +IYL+ +Y N +D + R V+AWH IR ++ QL +++ ++ SYTQW
Sbjct: 543 GYPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSSYTQW 722
Query: 353 VINRASDLVLPYHL 366
VI+RA + +PY +
Sbjct: 723 VIDRAVQIGMPYKI 764
>BE203370
Length = 454
Score = 274 bits (701), Expect = 5e-74
Identities = 131/151 (86%), Positives = 143/151 (93%)
Frame = +1
Query: 64 HCFTFPDYQLMPTLEEYSHLIGLPVLDKVPFTGLEPFPKAATIANALHLKTSLIKEKLTL 123
HCFTFPDYQL PTLEEYS+L+G PVLDKVPFTG EP PKAATIA+ALHL+TSLIK KLT+
Sbjct: 1 HCFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETSLIKAKLTI 180
Query: 124 KGNLPSLPTKFLYQQAFNFSKTNNVEAFYSILALLIYGLVLFPNIDNYVDIHAIQIFLTK 183
KGNLPSLPTKFLYQQA +F+K +NV+AFYSILALLIYGLVLFPN+DNYVDIHAIQ+FLTK
Sbjct: 181 KGNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLTK 360
Query: 184 NPVPTLLADIYHSIHDRTQVGRGAILGCAPL 214
NPVPTLLAD+YHSIHDR QVGRGAILGCAPL
Sbjct: 361 NPVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>BQ123351
Length = 725
Score = 105 bits (262), Expect = 4e-23
Identities = 52/140 (37%), Positives = 90/140 (64%)
Frame = +2
Query: 321 GRFVQAWHAIRTLNRSQLARRSDSLQGSYTQWVINRASDLVLPYHLPRYLSSTTPAPALP 380
G+F+QAW A+ + QL RRS + SYT+WVI+RA+ +P+ L R SSTT +P+LP
Sbjct: 2 GKFIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLP 181
Query: 381 LAPATVEECQERLAQSECVGATWKRKYDEAMLKMETMSGEIEQQEHEVHKLRRQIVKKNV 440
+ T EE Q LA+ TW+ +Y EA ++ T+ G++EQ++HE+ K+R+Q+++++
Sbjct: 182 MTSKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDD 361
Query: 441 QIRAQSTRLSQFISAGERWE 460
++ + L + I+ +R +
Sbjct: 362 LLQEKDKLLKKHITKKQRMD 421
>AL366606
Length = 393
Score = 81.3 bits (199), Expect = 8e-16
Identities = 42/78 (53%), Positives = 54/78 (68%), Gaps = 6/78 (7%)
Frame = -1
Query: 153 SILALLIYGLVLFPNIDNYVDIHAIQIFLTKNPVPTLLADIYHSIHDRTQVGRGAILGCA 212
+I L GL+LFPN+DN+VD++AI++F +KNPVPTLLAD YH+IHDRT GRG IL
Sbjct: 381 AIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYILWLH 202
Query: 213 PLL------YKWFTSHLP 224
L ++ F S LP
Sbjct: 201 ILFSNRVVYFRTFRSSLP 148
>AW736370
Length = 488
Score = 50.8 bits (120), Expect = 1e-06
Identities = 32/99 (32%), Positives = 50/99 (50%), Gaps = 1/99 (1%)
Frame = -2
Query: 374 TPAPALPLAPATVEECQERLAQSECVGATWKRKYDEAMLKMETMSGEIEQQEHEVHKLRR 433
TPAP LP+ T +E QERL ++E WKR+Y + ET+ G +EQ+ ++
Sbjct: 481 TPAPPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*KDV 302
Query: 434 QIVKKNVQIRAQSTRLSQFISAGE-RWEFFKDAHSDSDE 471
I K N +I+ L + + R + F HSD ++
Sbjct: 301 IIAKLNERIKENDAALDRIPGRKKRRMDLFDGPHSDFED 185
>BI269224 weakly similar to GP|11862972|db hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 537
Score = 31.2 bits (69), Expect = 0.90
Identities = 16/49 (32%), Positives = 27/49 (54%)
Frame = -3
Query: 406 KYDEAMLKMETMSGEIEQQEHEVHKLRRQIVKKNVQIRAQSTRLSQFIS 454
KY E M + S + + +V+ ++VKKN ++ QST+ S+ IS
Sbjct: 490 KYTEVMTSR*SCSRQYITNDDDVNNKNTKVVKKNRNLKKQSTKTSKSIS 344
>AL388248 weakly similar to GP|6062758|gb|A NADH dehydrogenase subunit II
{Cynolebias alexandri}, partial (7%)
Length = 417
Score = 29.6 bits (65), Expect = 2.6
Identities = 15/59 (25%), Positives = 30/59 (50%)
Frame = -2
Query: 385 TVEECQERLAQSECVGATWKRKYDEAMLKMETMSGEIEQQEHEVHKLRRQIVKKNVQIR 443
T+++ ERL + A WK KY+ + + E GE V +L ++++ + Q++
Sbjct: 302 TLQKHNERLKEKL---ANWKEKYEASETEREAAEGEASAANASVRRLTMKVLELSKQLK 135
>TC80272 similar to GP|15620810|dbj|BAB67768. RPS6-like protein {Arabidopsis
thaliana}, partial (35%)
Length = 1026
Score = 28.9 bits (63), Expect = 4.4
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 7/61 (11%)
Frame = -1
Query: 137 QQAFNFSKTNNVEAFYSILALLIYGLVLFPN-------IDNYVDIHAIQIFLTKNPVPTL 189
++ F KTN +SIL +L G ++FPN ++ H QI L + P L
Sbjct: 930 EKKFKIKKTNTYLPTFSILIMLFSGPLIFPNHHRL**KFHHHPQYHHNQIHLPRLHHPIL 751
Query: 190 L 190
L
Sbjct: 750 L 748
>TC86966 similar to GP|6850934|emb|CAB71030.1 lectin-like protein {Cicer
arietinum}, partial (82%)
Length = 942
Score = 28.9 bits (63), Expect = 4.4
Identities = 19/73 (26%), Positives = 32/73 (43%)
Frame = -3
Query: 158 LIYGLVLFPNIDNYVDIHAIQIFLTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYK 217
LI+ LFPN ++ ++ +P T+L + +H H C P LY
Sbjct: 385 LIFI*SLFPNFSHHK-------YICASPTETILLNFHHKFH----------YVCIPFLYF 257
Query: 218 WFTSHLPQTHSFQ 230
+FT +L + +Q
Sbjct: 256 YFTRYLQP*NLYQ 218
>TC79953 similar to GP|20279471|gb|AAM18751.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (25%)
Length = 899
Score = 28.9 bits (63), Expect = 4.4
Identities = 12/37 (32%), Positives = 23/37 (61%)
Frame = -3
Query: 423 QQEHEVHKLRRQIVKKNVQIRAQSTRLSQFISAGERW 459
+Q H RR+IV + + IR S+R+++ +++ RW
Sbjct: 498 RQRHRNSSRRRRIVIRIIMIRRSSSRVNKSVTSSLRW 388
>TC88618 weakly similar to PIR|T48397|T48397 S-receptor kinase-like protein -
Arabidopsis thaliana, partial (38%)
Length = 2304
Score = 28.5 bits (62), Expect = 5.8
Identities = 27/123 (21%), Positives = 47/123 (37%), Gaps = 9/123 (7%)
Frame = -3
Query: 54 TLVQFYDPAYHCFTFPDYQLMPTLEEYSH---------LIGLPVLDKVPFTGLEPFPKAA 104
TL Y+P ++ F Y +P + +SH + LP++ L+PF ++
Sbjct: 1168 TLFLLYNPKHNKTRFGYYTKLPNIYNFSHTNPHILLKITLLLPLVTLYAVAPLQPFSESC 989
Query: 105 TIANALHLKTSLIKEKLTLKGNLPSLPTKFLYQQAFNFSKTNNVEAFYSILALLIYGLVL 164
+LH L K ++ + PT+F T + +I I+ L
Sbjct: 988 NY--SLHQSLPLHKSPTSITIIIIKTPTRFKPSFVI*TRTTRPTRSTQTIRTTWIWKLAS 815
Query: 165 FPN 167
F N
Sbjct: 814 FTN 806
>TC90790 similar to GP|18086412|gb|AAL57663.1 At2g17250/T23A1.11
{Arabidopsis thaliana}, partial (23%)
Length = 812
Score = 28.5 bits (62), Expect = 5.8
Identities = 15/34 (44%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Frame = +1
Query: 377 PALPLAPATVEECQERLAQSECVGATWKR-KYDE 409
P LP + AT QSEC+ TW R K+DE
Sbjct: 232 PQLPSSAATSSSSSADSDQSECIYLTWLRSKFDE 333
>AW775436
Length = 659
Score = 28.1 bits (61), Expect = 7.6
Identities = 17/62 (27%), Positives = 32/62 (51%)
Frame = -1
Query: 118 KEKLTLKGNLPSLPTKFLYQQAFNFSKTNNVEAFYSILALLIYGLVLFPNIDNYVDIHAI 177
+E K + PSL LY +F F ++ +F++IL G+++FP ++ H++
Sbjct: 584 RENKCSKPSNPSLS*NMLYIVSFKFPCHSHSSSFFAILTF*RSGVLVFPAEKIFMRGHSL 405
Query: 178 QI 179
I
Sbjct: 404 TI 399
>BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvox carteri,
partial (9%)
Length = 1337
Score = 28.1 bits (61), Expect = 7.6
Identities = 16/44 (36%), Positives = 19/44 (42%)
Frame = -3
Query: 87 PVLDKVPFTGLEPFPKAATIANALHLKTSLIKEKLTLKGNLPSL 130
PVL + PFP +A I A HL+ L LPSL
Sbjct: 903 PVLSRSLILTYPPFPASAVILRASHLRYRFAPSYLRSSAPLPSL 772
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,114,289
Number of Sequences: 36976
Number of extensions: 255746
Number of successful extensions: 1246
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1238
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1245
length of query: 471
length of database: 9,014,727
effective HSP length: 100
effective length of query: 371
effective length of database: 5,317,127
effective search space: 1972654117
effective search space used: 1972654117
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144345.13


