

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.7 + phase: 0
(745 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
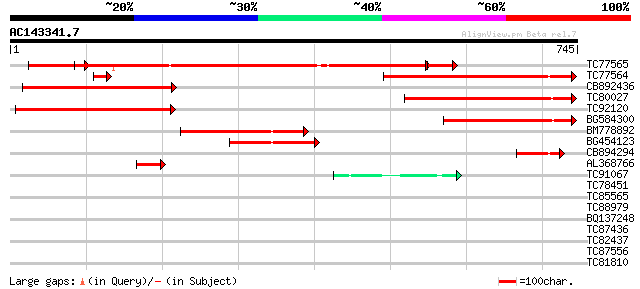
Score E
Sequences producing significant alignments: (bits) Value
TC77565 similar to GP|21592665|gb|AAM64614.1 Isp4-like protein {... 509 e-158
TC77564 similar to GP|20466754|gb|AAM20694.1 Isp4-like protein {... 341 7e-94
CB892436 similar to PIR|T08925|T08 isp4 protein homolog T15N24.4... 338 4e-93
TC80027 similar to GP|15451020|gb|AAK96781.1 Unknown protein {Ar... 270 1e-72
TC92120 similar to PIR|T01900|T01900 isp4 protein homolog T12H20... 242 4e-64
BG584300 similar to GP|6983868|dbj EST D25093(R3139) corresponds... 216 3e-56
BM778892 similar to GP|6983868|dbj EST D25093(R3139) corresponds... 178 8e-45
BG454123 similar to PIR|T01900|T01 isp4 protein homolog T12H20.7... 107 2e-23
CB894294 similar to GP|20160527|dbj putative sexual differentiat... 80 2e-15
AL368766 GP|20466754|gb Isp4-like protein {Arabidopsis thaliana}... 57 2e-08
TC91067 similar to PIR|H96681|H96681 protein F1E22.10 [imported]... 42 9e-04
TC78451 similar to GP|9279625|dbj|BAB01083.1 gb|AAF23830.1~gene_... 34 0.18
TC85565 homologue to SP|P27880|HS12_MEDSA 18.2 kDa class I heat ... 31 2.0
TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.... 31 2.0
BQ137248 30 2.6
TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyas... 30 2.6
TC82437 weakly similar to PIR|E71416|E71416 hypothetical protein... 30 3.4
TC87556 similar to GP|14517530|gb|AAK62655.1 At1g48370/F11A17_27... 30 4.4
TC81810 weakly similar to PIR|T04255|T04255 hypothetical protein... 29 5.8
>TC77565 similar to GP|21592665|gb|AAM64614.1 Isp4-like protein {Arabidopsis
thaliana}, partial (71%)
Length = 1698
Score = 509 bits (1310), Expect(2) = e-158
Identities = 243/475 (51%), Positives = 344/475 (72%), Gaps = 7/475 (1%)
Frame = +1
Query: 86 LPLGKLMAATLPTKPIQVP-FTTWSFSLNPGPFSLKEHVLITIFASSGSS----GVYAIN 140
LP+G MAA LPT ++P F T SLNPGPF++KEHVLITIFA++GS+ YA+
Sbjct: 193 LPIGHFMAAILPTTKFRIPGFGTKKLSLNPGPFNMKEHVLITIFANAGSAFGSGSPYAVG 372
Query: 141 IITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRA 200
I+ I+KAFY RSI AA+LL ++TQ+LGYGWAG+ R+++V+ +MWWP +LVQVSLFRA
Sbjct: 373 IVNIIKAFYGRSISFHAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPSTLVQVSLFRA 552
Query: 201 FHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGS 260
HEK+ R SR +FFF+ V SF++Y+IPGY F ++++S+VC ++ +S+TAQQ+GS
Sbjct: 553 LHEKDDDHR--ISRAKFFFIALVCSFSWYVIPGYLFTTLTSISWVCWVFTKSVTAQQLGS 726
Query: 261 GMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NLYDAK 319
GMKGLGIG+ LDW VA FL SPL P FAI+N+ G+ L +Y ++PI+YW N Y+AK
Sbjct: 727 GMKGLGIGAITLDWTAVASFLFSPLISPFFAIVNVFVGYVLIIYAVVPIAYWGLNAYNAK 906
Query: 320 KFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTA 379
FPL SSH F + G YN++ I+N + F++D Y+ +I+LSV F+ YGF FA + +
Sbjct: 907 TFPLFSSHLFTAQGQKYNISAIVN-DKFELDEAKYAEQGRIHLSVFFSLTYGFGFATIAS 1083
Query: 380 TISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMAL 439
T++HV F+G I++ ++ ++ + D+HT++MK+ Y+ +P WWF +L++ + ++L
Sbjct: 1084TLTHVACFYGREIMERYRASSKGKE----DIHTKLMKR-YKDIPSWWFYALLVVTLAVSL 1248
Query: 440 LACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKP 499
+ C Q+Q+PWWG+L + +A FTLPI +I ATTN GLN+ITE V G IYPG+P
Sbjct: 1249VLCIFLNDQIQMPWWGLLFAGALAFAFTLPISIITATTNQTPGLNIITEYVFGLIYPGRP 1428
Query: 500 LANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVA-SSVHFG 553
+ANV FKTYG+ISM QA+ FL DFKLGHYMKI P+SMF+VQ +GT++A + H+G
Sbjct: 1429IANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTMLARNHQHWG 1593
Score = 84.7 bits (208), Expect = 1e-16
Identities = 42/80 (52%), Positives = 50/80 (62%)
Frame = +3
Query: 25 SPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQII 84
SPIE+VRLTV TDDP+QP TFR W LGL SC LL+F+NQF YRT PL IT ++ Q+
Sbjct: 9 SPIEEVRLTVTNTDDPTQPVWTFRMWFLGLISCSLLSFLNQFFAYRTEPLIITLITVQVA 188
Query: 85 ALPLGKLMAATLPTKPIQVP 104
P L +Q P
Sbjct: 189 YTPYRSLHGGYSANHEVQNP 248
Score = 68.6 bits (166), Expect(2) = e-158
Identities = 25/40 (62%), Positives = 33/40 (82%)
Frame = +2
Query: 549 SVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNA 588
+++ G AWWLL S+ENIC + LLP+GSPWTCPGD F++A
Sbjct: 1577 TINIGVAWWLLNSVENICHDDLLPEGSPWTCPGDPCFFDA 1696
>TC77564 similar to GP|20466754|gb|AAM20694.1 Isp4-like protein {Arabidopsis
thaliana}, partial (37%)
Length = 1275
Score = 341 bits (874), Expect = 7e-94
Identities = 148/253 (58%), Positives = 189/253 (74%)
Frame = +1
Query: 492 GFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVH 551
G IYPG+P+ANV FKTYG+ISM QA+ FL DFKLGHYMKI P+SMF+VQ +GT++A +++
Sbjct: 79 GLIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTMLAGTIN 258
Query: 552 FGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELN 611
G AWWLL S+ENIC + LLP+GSPWTCPGD VF++AS+IWG+VGPKR+F G Y +N
Sbjct: 259 IGVAWWLLNSVENICHDDLLPEGSPWTCPGDRVFFDASVIWGLVGPKRIFGSKGNYSAMN 438
Query: 612 WFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFN 671
WFF+ G I P+ VWLL FP Q WI IN+P+++ + +PP +NY +W VG FN
Sbjct: 439 WFFIGGAIGPIIVWLLHKAFPRQSWIALINLPVLLGATAMMPPATPLNYNSWIFVGTIFN 618
Query: 672 FYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKC 731
F+V+R K WW R+ Y+LSA LD GVAF+ +LLYFSL S +WWG E +HCPLA C
Sbjct: 619 FFVFRYRKKWWQRYNYVLSAALDTGVAFMTVLLYFSL-SLENRSISWWGTEGEHCPLASC 795
Query: 732 PTAPGVHAEGCPV 744
PTA G+ +GCPV
Sbjct: 796 PTAKGIAVDGCPV 834
Score = 45.1 bits (105), Expect = 1e-04
Identities = 19/24 (79%), Positives = 24/24 (99%)
Frame = +1
Query: 111 SLNPGPFSLKEHVLITIFASSGSS 134
SLNPGPF++KEHVLITIFA++GS+
Sbjct: 4 SLNPGPFNMKEHVLITIFANAGSA 75
>CB892436 similar to PIR|T08925|T08 isp4 protein homolog T15N24.40 -
Arabidopsis thaliana, partial (24%)
Length = 707
Score = 338 bits (867), Expect = 4e-93
Identities = 159/202 (78%), Positives = 185/202 (90%)
Frame = +2
Query: 18 DEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKIT 77
D+ ++DSPIEQVRLTV TDDP+QPALTFRTW++GLA CI+LAFVNQF GYRTNPL IT
Sbjct: 95 DDTEVDDSPIEQVRLTVSTTDDPTQPALTFRTWIIGLACCIVLAFVNQFFGYRTNPLTIT 274
Query: 78 SVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVY 137
+VSAQI++LP+GKLMAATLPT +VPFT WSF+LNPGPF+LKEH LITIFAS+G+ GVY
Sbjct: 275 AVSAQIVSLPIGKLMAATLPTTIYKVPFTKWSFTLNPGPFNLKEHALITIFASAGAGGVY 454
Query: 138 AINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSL 197
AINIITIVKAFYHR+I+P+AA+LLA++TQMLGYGWAG+FRRFLVDSPYMWWP +LVQVSL
Sbjct: 455 AINIITIVKAFYHRNINPIAAFLLAITTQMLGYGWAGMFRRFLVDSPYMWWPFNLVQVSL 634
Query: 198 FRAFHEKEKRPRGGTSRLQFFF 219
FRAFHEKEKR +G T+RLQF F
Sbjct: 635 FRAFHEKEKRLKGRTTRLQFLF 700
>TC80027 similar to GP|15451020|gb|AAK96781.1 Unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 926
Score = 270 bits (690), Expect = 1e-72
Identities = 117/226 (51%), Positives = 159/226 (69%)
Frame = +1
Query: 519 FLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENICDESLLPKGSPWT 578
FL + KLGHYMKI P+ M+ QLVGT+VA V+ AWW+L SI++IC + SPWT
Sbjct: 4 FLQNLKLGHYMKIPPRCMYTAQLVGTLVAGVVNLSVAWWMLDSIKDICMDDKAHHDSPWT 183
Query: 579 CPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQ 638
CP V ++AS+IWG++GPKR+F G+Y L W FL+G + PVPVW+ S +PN+KWI
Sbjct: 184 CPKYRVTFDASVIWGLIGPKRLFGPGGLYRNLVWLFLVGAVLPVPVWVFSKIYPNKKWIP 363
Query: 639 FINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVA 698
INIP+I G + +PP N +W + G+ FN++V+R K WW ++ Y+LSA LDAG A
Sbjct: 364 LINIPVISYGFAGMPPATPTNIASWLLTGMIFNYFVFRFHKRWWQKYNYVLSAALDAGTA 543
Query: 699 FIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVHAEGCPV 744
F+G+L++F+LQ+ G + WWG E DHCPLA CPTAPG+ +GCPV
Sbjct: 544 FMGVLIFFALQNAG-HTLKWWGTELDHCPLATCPTAPGIVVDGCPV 678
>TC92120 similar to PIR|T01900|T01900 isp4 protein homolog T12H20.7 -
Arabidopsis thaliana, partial (25%)
Length = 647
Score = 242 bits (617), Expect = 4e-64
Identities = 114/211 (54%), Positives = 157/211 (74%)
Frame = +2
Query: 8 QNTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFL 67
+ +I++++ + +SPIEQV LTVP+TDDPS P TFRTW LG +C+LL+F+NQF
Sbjct: 17 KEALIKNSDATRSNGENSPIEQVALTVPVTDDPSLPVFTFRTWTLGTLACVLLSFLNQFF 196
Query: 68 GYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITI 127
G+R PL +T++SAQI +PLG LMA+T+ TK + + W F+LNPG F++KEHVLITI
Sbjct: 197 GFRREPLSVTAISAQIAVVPLGHLMASTI-TKRVFMKGKKWEFTLNPGKFNVKEHVLITI 373
Query: 128 FASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMW 187
FASSG++ VYAI+ ++ VK FY + I + A L+ L+TQ+LG+GWAG+F R+LV+ MW
Sbjct: 374 FASSGAASVYAIHFVSTVKVFYRKEITVLVALLVVLTTQVLGFGWAGVFXRYLVEPAGMW 553
Query: 188 WPESLVQVSLFRAFHEKEKRPRGGTSRLQFF 218
WP++LVQVSLFRA KE R +GG + QFF
Sbjct: 554 WPQNLVQVSLFRALXXKEXRQKGGLTXNQFF 646
>BG584300 similar to GP|6983868|dbj EST D25093(R3139) corresponds to a region
of the predicted gene.~Similar to Arabidopsis thaliana,
partial (21%)
Length = 755
Score = 216 bits (549), Expect = 3e-56
Identities = 88/175 (50%), Positives = 122/175 (69%)
Frame = +3
Query: 570 LLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIAPVPVWLLSL 629
+LP GSPWTCPGD VFY++S++WG++GP+R+F G Y +NWFFL+G +AP VWL
Sbjct: 27 VLPAGSPWTCPGDHVFYDSSVVWGLIGPRRIFGNLGHYSAINWFFLVGAVAPFIVWLAHK 206
Query: 630 KFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKAWWARHTYIL 689
P+++WI+ I +P+I+ +++PP VNY +W +VG F YR ++ WW RH Y+L
Sbjct: 207 ALPDKQWIKLITMPVILGALTEMPPATPVNYTSWVLVGFASGFVAYRYYRGWWTRHNYVL 386
Query: 690 SAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVHAEGCPV 744
S LDAG+AF+G+L+Y L I WWG +PD CPLA CPTAPGV + GCP+
Sbjct: 387 SGALDAGLAFMGVLIYLCLGMQHI-SLDWWGSDPDRCPLASCPTAPGVISAGCPL 548
>BM778892 similar to GP|6983868|dbj EST D25093(R3139) corresponds to a region
of the predicted gene.~Similar to Arabidopsis thaliana,
partial (22%)
Length = 852
Score = 178 bits (451), Expect = 8e-45
Identities = 77/168 (45%), Positives = 118/168 (69%)
Frame = +1
Query: 225 SFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSP 284
SFAYY++PGY F ++++S++C ++ SI AQQ+GSG+ GLG+G+ G DW+++ +LGSP
Sbjct: 1 SFAYYVLPGYLFPMLTSLSWICWVFPNSIIAQQLGSGLHGLGVGAIGFDWSSICSYLGSP 180
Query: 285 LAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNT 344
LA P FA NI AGF ++MYV++PI+Y NLY + FP+ S F++ G YN++ I+++
Sbjct: 181 LASPWFATANIAAGFGIFMYVVVPIAYGLNLYHGRSFPIFSDGLFNTNGQEYNISAIIDS 360
Query: 345 ETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTATISHVVLFHGEMI 392
F +D+++Y +YLS FA YG FA L+A + HV+LFHG +
Sbjct: 361 -NFHLDLDAYQREGPLYLSTMFAMSYGIDFACLSAILVHVLLFHGRYV 501
>BG454123 similar to PIR|T01900|T01 isp4 protein homolog T12H20.7 -
Arabidopsis thaliana, partial (13%)
Length = 654
Score = 107 bits (267), Expect = 2e-23
Identities = 51/118 (43%), Positives = 74/118 (62%)
Frame = +1
Query: 290 FAIINIMAGFFLYMYVLIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDI 349
FA N+ GF MY+L P+ YW N+Y+AK FP+ S+ F S G YN+T I+++ F +
Sbjct: 1 FATANVAVGFVFVMYILTPLCYWFNVYNAKTFPIFSNQLFTSKGEIYNITEIIDS-NFHM 177
Query: 350 DMESYSNYSKIYLSVAFAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQL 407
D+ +Y +++LS FA YG FAALTATI HV LFHG ++ + +T++ N L
Sbjct: 178 DLAAYEKQGRLHLSTFFAMTYGVGFAALTATIVHVALFHGRYVLFVSTLNSTNIFNVL 351
>CB894294 similar to GP|20160527|dbj putative sexual differentiation process
protein Isp4 {Oryza sativa (japonica cultivar-group)},
partial (8%)
Length = 190
Score = 80.5 bits (197), Expect = 2e-15
Identities = 35/63 (55%), Positives = 47/63 (74%)
Frame = +2
Query: 666 VGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDH 725
VG F+F+V+R + WW R++Y+LSA LD GVAF+ +LLYFSL S + G +WWG + DH
Sbjct: 5 VGTIFSFFVFRYRRRWWQRYSYVLSAALDTGVAFMTVLLYFSL-SLELRGISWWGADGDH 181
Query: 726 CPL 728
CPL
Sbjct: 182 CPL 190
>AL368766 GP|20466754|gb Isp4-like protein {Arabidopsis thaliana}, partial
(4%)
Length = 516
Score = 57.4 bits (137), Expect = 2e-08
Identities = 22/38 (57%), Positives = 32/38 (83%)
Frame = -1
Query: 167 MLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAFHEK 204
+LGYGWAG+ R+++V+ +MWWP +LVQVSLFR ++K
Sbjct: 114 VLGYGWAGLLRKYVVEPAHMWWPSTLVQVSLFRYDNKK 1
>TC91067 similar to PIR|H96681|H96681 protein F1E22.10 [imported] -
Arabidopsis thaliana, partial (46%)
Length = 1223
Score = 42.0 bits (97), Expect = 9e-04
Identities = 41/168 (24%), Positives = 66/168 (38%)
Frame = +3
Query: 426 WFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNV 485
W+ ILI ++ AL C +G L W + + IFT+ A + +GL
Sbjct: 345 WYHVILIYIIAPALAFCNAYG--CGLTDWSLASTYGKLAIFTIGAWAGSANGGVLAGLAA 518
Query: 486 ITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTV 545
++ N+ + A + DFK G+ +PKSMF+ Q++GT
Sbjct: 519 CGVMM-----------NI---------VSTASDLMQDFKTGYMTLASPKSMFVSQVIGTA 638
Query: 546 VASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWG 593
+ + W + +L GS + P VF N +II G
Sbjct: 639 MGCIISPCVFWLFYHAF-----GTLGQPGSAYPAPYALVFRNIAIIGG 767
>TC78451 similar to GP|9279625|dbj|BAB01083.1
gb|AAF23830.1~gene_id:MOJ10.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (88%)
Length = 2452
Score = 34.3 bits (77), Expect = 0.18
Identities = 37/151 (24%), Positives = 60/151 (39%)
Frame = +2
Query: 497 GKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAW 556
G +A VA + A + DFK G+ + KSMF+ QL+GT + + T W
Sbjct: 1568 GGVIAGVASCAVMMSIVATAADLMQDFKTGYLTLSSAKSMFVSQLIGTAMGCIIAPLTFW 1747
Query: 557 WLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLI 616
T+ + + P+ P +F +I+ GV G + P+
Sbjct: 1748 MFWTAFD------IGSPDGPYKAPYAVIFREMAIL-GVEGFSEL-------PKYCMEMCG 1885
Query: 617 GLIAPVPVWLLSLKFPNQKWIQFINIPIIIA 647
G A L +K+ Q+I IP+ +A
Sbjct: 1886 GFFAAALAINLLRDVTPKKYSQYIPIPMAMA 1978
>TC85565 homologue to SP|P27880|HS12_MEDSA 18.2 kDa class I heat shock
protein. [Alfalfa] {Medicago sativa}, complete
Length = 735
Score = 30.8 bits (68), Expect = 2.0
Identities = 22/75 (29%), Positives = 36/75 (47%)
Frame = +2
Query: 161 LALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFFFV 220
+ALSTQ++ + + + +VDS ++WWP+ FR+ P G S QF
Sbjct: 32 IALSTQLVLSLQSSLLLQNVVDSKFLWWPKE----QRFRSILP*RLGPLQGFSFNQF--- 190
Query: 221 VFVASFAYYIIPGYF 235
+F ++I G F
Sbjct: 191 ---CTFCFFIPSGEF 226
>TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.14
[imported] - Arabidopsis thaliana, partial (89%)
Length = 1616
Score = 30.8 bits (68), Expect = 2.0
Identities = 16/38 (42%), Positives = 22/38 (57%)
Frame = -1
Query: 559 LTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVG 596
+ S ICD++LLP+ SP TC Y AS I+ + G
Sbjct: 500 MESTSRICDKNLLPRPSP*TC----TLYKASNIYKLYG 399
>BQ137248
Length = 977
Score = 30.4 bits (67), Expect = 2.6
Identities = 23/75 (30%), Positives = 34/75 (44%), Gaps = 3/75 (4%)
Frame = -3
Query: 235 FFQAISTVSFVCLIWKESITAQQIGSGMKGLGIGSFGLDWNTV---AGFLGSPLAVPGFA 291
FF +S + +VC + ++ + + G+G G FGL +TV GF + G
Sbjct: 300 FFSVLSLLFYVCFRFHFFFSSVTLL*SVLGVGAGGFGLSTDTVRCGCGFRFRSASFDG-- 127
Query: 292 IINIMAGFFLYMYVL 306
GF L MYVL
Sbjct: 126 -----PGFCLVMYVL 97
>TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyase-like
protein - Arabidopsis thaliana, partial (75%)
Length = 1592
Score = 30.4 bits (67), Expect = 2.6
Identities = 18/62 (29%), Positives = 34/62 (54%), Gaps = 8/62 (12%)
Frame = +2
Query: 412 TRIMKKNYEQVPE-WWFVTILILMVMMALLACEGFGKQLQLPW-------WGILLSLTIA 463
+ ++ +N E+V WWF+ +LIL +++ L F + L L W WG++L + +
Sbjct: 1319 SEMLFRNLEEVRLLWWFLKVLILWMLVDLC---WFRRSLGLGWMLVLGGLWGLVLVIVLR 1489
Query: 464 LI 465
L+
Sbjct: 1490 LL 1495
>TC82437 weakly similar to PIR|E71416|E71416 hypothetical protein -
Arabidopsis thaliana, partial (42%)
Length = 1680
Score = 30.0 bits (66), Expect = 3.4
Identities = 21/60 (35%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Frame = +2
Query: 410 VHT-RIMKKNYEQVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTL 468
VHT R M + V EW V +++L+V + L + F L L W G L+ + + + FTL
Sbjct: 941 VHTLRFMSRIVCLVLEWLLVVLVLLLVHLLLKFWQRF---LILVWKGTLIYMEVMVEFTL 1111
>TC87556 similar to GP|14517530|gb|AAK62655.1 At1g48370/F11A17_27
{Arabidopsis thaliana}, partial (48%)
Length = 1309
Score = 29.6 bits (65), Expect = 4.4
Identities = 12/41 (29%), Positives = 22/41 (53%)
Frame = +1
Query: 516 ALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAW 556
A + DFK G+ +P+S+F+ Q++GT + + W
Sbjct: 526 ASDLMQDFKTGYLTLASPRSVFVSQIIGTTMGCIISPCVFW 648
>TC81810 weakly similar to PIR|T04255|T04255 hypothetical protein F20B18.110
- Arabidopsis thaliana, partial (12%)
Length = 801
Score = 29.3 bits (64), Expect = 5.8
Identities = 22/67 (32%), Positives = 29/67 (42%), Gaps = 3/67 (4%)
Frame = +1
Query: 209 RGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSG---MKGL 265
+ G F F VF+ S YI FF I+T+ + IW + Q G +KGL
Sbjct: 208 KNGCGSKVFIFSVFILSVGQYITYPAFFIVITTIHRLWSIWFRRL*YFQALVGISLVKGL 387
Query: 266 GIGSFGL 272
FGL
Sbjct: 388 KCTPFGL 408
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.141 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,309,816
Number of Sequences: 36976
Number of extensions: 418996
Number of successful extensions: 2987
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 2165
Number of HSP's successfully gapped in prelim test: 124
Number of HSP's that attempted gapping in prelim test: 733
Number of HSP's gapped (non-prelim): 2382
length of query: 745
length of database: 9,014,727
effective HSP length: 103
effective length of query: 642
effective length of database: 5,206,199
effective search space: 3342379758
effective search space used: 3342379758
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC143341.7


