

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.10 + phase: 0
(633 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
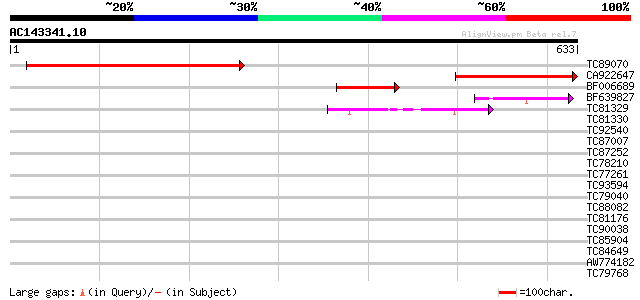
Score E
Sequences producing significant alignments: (bits) Value
TC89070 similar to GP|20197343|gb|AAM15033.1 putative helicase {... 471 e-133
CA922647 similar to GP|11191231|em putative helicase {Arabidopsi... 266 2e-71
BF006689 similar to GP|9758800|dbj| DNA helicase-like {Arabidops... 68 1e-11
BF639827 similar to PIR|T00740|T007 hypothetical protein F22O13.... 66 4e-11
TC81329 homologue to GP|20466796|gb|AAM20715.1 unknown protein {... 47 2e-05
TC81330 homologue to GP|20466796|gb|AAM20715.1 unknown protein {... 35 0.088
TC92540 similar to GP|22136924|gb|AAM91806.1 putative RNA helica... 34 0.20
TC87007 homologue to PIR|T48023|T48023 ATP-dependent RNA helicas... 33 0.43
TC87252 similar to GP|11559424|dbj|BAB18781. mitochondrial prote... 32 0.74
TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Ar... 32 0.74
TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AA... 32 0.74
TC93594 weakly similar to GP|20466452|gb|AAM20543.1 unknown prot... 32 0.74
TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase ... 32 0.97
TC88082 similar to GP|10798790|dbj|BAB16439. replication factor ... 31 1.3
TC81176 similar to GP|17065122|gb|AAL32715.1 Similar replication... 31 1.7
TC90038 similar to PIR|A84451|A84451 probable AAA-type ATPase [i... 30 2.2
TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [i... 30 2.2
TC84649 similar to PIR|A84451|A84451 probable AAA-type ATPase [i... 30 2.2
AW774182 homologue to PIR|F84674|F84 probable AAA-type ATPase [i... 30 2.8
TC79768 similar to PIR|B96657|B96657 probable replication factor... 30 3.7
>TC89070 similar to GP|20197343|gb|AAM15033.1 putative helicase {Arabidopsis
thaliana}, partial (38%)
Length = 861
Score = 471 bits (1213), Expect = e-133
Identities = 243/244 (99%), Positives = 244/244 (99%)
Frame = +2
Query: 19 EISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKADVLPAHKFGTH 78
EISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKADVLPAHKFGTH
Sbjct: 128 EISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKADVLPAHKFGTH 307
Query: 79 DVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLRLEKVANEVTYHRM 138
DVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLRLEKVANEVTYHRM
Sbjct: 308 DVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLRLEKVANEVTYHRM 487
Query: 139 KDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDYSQKDAISKALSSK 198
KDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDYSQKDAISKALSSK
Sbjct: 488 KDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDYSQKDAISKALSSK 667
Query: 199 NVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRIGH 258
NVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLV+IGH
Sbjct: 668 NVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVKIGH 847
Query: 259 PARL 262
PARL
Sbjct: 848 PARL 859
>CA922647 similar to GP|11191231|em putative helicase {Arabidopsis thaliana},
partial (21%)
Length = 711
Score = 266 bits (680), Expect = 2e-71
Identities = 136/136 (100%), Positives = 136/136 (100%)
Frame = -3
Query: 498 EGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQG 557
EGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQG
Sbjct: 709 EGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQG 530
Query: 558 REKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEY 617
REKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEY
Sbjct: 529 REKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEY 350
Query: 618 FEEHGEYQSASEYQNE 633
FEEHGEYQSASEYQNE
Sbjct: 349 FEEHGEYQSASEYQNE 302
>BF006689 similar to GP|9758800|dbj| DNA helicase-like {Arabidopsis
thaliana}, partial (9%)
Length = 214
Score = 67.8 bits (164), Expect = 1e-11
Identities = 35/70 (50%), Positives = 46/70 (65%)
Frame = +2
Query: 366 QALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSML 425
QA+E +CWIP+L+ RCILAGD QL P I S +A + G +L ER A L+ +T+ L
Sbjct: 2 QAIEPSCWIPILQAKRCILAGDQCQLAPVIFSRKALEGGSRISLLERAATLHEGILTTRL 181
Query: 426 TVQYRMHQLI 435
T QYRM+ I
Sbjct: 182 TTQYRMNDAI 211
>BF639827 similar to PIR|T00740|T007 hypothetical protein F22O13.35 -
Arabidopsis thaliana, partial (8%)
Length = 524
Score = 66.2 bits (160), Expect = 4e-11
Identities = 41/113 (36%), Positives = 63/113 (55%), Gaps = 3/113 (2%)
Frame = +1
Query: 520 DIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKE---V 576
+IGIITPY +Q L++ + +EI T+D +QGR+K+ I++S VRS
Sbjct: 10 NIGIITPYNSQANLIR----HATCITSLEIHTIDKYQGRDKDCILVSFVRSCENPTSCVA 177
Query: 577 GFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEEHGEYQSASE 629
L D R+NVA+TRA+R+ +V +T+ LK LI+ EE + S+
Sbjct: 178 SLLGDWHRINVALTRAKRKLIMVGSRKTLLKVPLLKLLIKKVEEQSGILNVSK 336
>TC81329 homologue to GP|20466796|gb|AAM20715.1 unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 963
Score = 47.4 bits (111), Expect = 2e-05
Identities = 52/203 (25%), Positives = 94/203 (45%), Gaps = 18/203 (8%)
Frame = +1
Query: 356 FDLVIIDEAAQALEVACWIPLL---------KGTRCILAGDHLQLPPTIQSVEAEK-KGL 405
+D ++++E+AQ LE+ +IP+L + RCIL GDH QLPP ++++ +K +
Sbjct: 373 YDNLLMEESAQILEIETFIPMLLQRQEDGHARLKRCILIGDHHQLPPVVKNMAFQKYSHM 552
Query: 406 GRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGV 465
++LF R L + L Q R S +LYN + + L DL +
Sbjct: 553 DQSLFTRFVRLGIPYIE--LNAQGRARP-----SIAKLYNWRYRD--------LGDLPFL 687
Query: 466 KKTS--STEPTLLLIDTAGCDMEEKKDEEDST------LNEGESEVAMAHAKRLVQSGVL 517
K+ + + D D+ + + ++T NEGE+E ++ + G
Sbjct: 688 KEEAIFNRANAGFAYDYQLVDVPDHNGKGETTPSPWFYQNEGEAEYIVSVYIYMRLLGYP 867
Query: 518 PSDIGIITPYAAQVVLLKMLKNK 540
+ I I+T Y Q +L++ + N+
Sbjct: 868 ANKISILTTYNGQKLLIRDVINR 936
>TC81330 homologue to GP|20466796|gb|AAM20715.1 unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 698
Score = 35.0 bits (79), Expect = 0.088
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 5/76 (6%)
Frame = +3
Query: 205 GPPGTGKTTTVVEI--ILQEVKRGSKILACAASNIAVDNIVERLVPHRVK---LVRIGHP 259
GPPGTGKT T V+I +L + L SN A++++ E+++ V L+R+G
Sbjct: 198 GPPGTGKTDTAVQILNVLYHNCPSQRTLIITHSNQALNDLFEKIMQRDVPARYLLRLGQG 377
Query: 260 ARLLPQVVDSALDAQV 275
+ L +D + +V
Sbjct: 378 EQELATDLDFSRQGRV 425
>TC92540 similar to GP|22136924|gb|AAM91806.1 putative RNA helicase
{Arabidopsis thaliana}, partial (24%)
Length = 836
Score = 33.9 bits (76), Expect = 0.20
Identities = 22/99 (22%), Positives = 43/99 (43%)
Frame = +1
Query: 185 YSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVE 244
Y+ +D + +A+ V ++ G G+GKTT + + + + ++AC V
Sbjct: 373 YTYRDELLQAVHDHQVLVIVGETGSGKTTQIPQYLHEAGYTKHGMIACTQPRRVAAMSVA 552
Query: 245 RLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGL 283
V H + V++GH + D D +L+ G+
Sbjct: 553 ARVAHELG-VKLGHEVGYSIRFEDCTSDKTILKYMTDGM 666
>TC87007 homologue to PIR|T48023|T48023 ATP-dependent RNA helicase-like
protein - Arabidopsis thaliana, partial (96%)
Length = 2361
Score = 32.7 bits (73), Expect = 0.43
Identities = 17/54 (31%), Positives = 27/54 (49%), Gaps = 6/54 (11%)
Frame = +3
Query: 185 YSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEV------KRGSKILAC 232
+ QK+ K L +L G G+GKTT + + +L+ V KR ++AC
Sbjct: 351 WHQKEDFLKVLKDNQTLILVGETGSGKTTQIPQFVLEAVELEAPDKRKKMMIAC 512
>TC87252 similar to GP|11559424|dbj|BAB18781. mitochondrial protein-like
protein {Cucumis sativus}, partial (72%)
Length = 968
Score = 32.0 bits (71), Expect = 0.74
Identities = 22/63 (34%), Positives = 30/63 (46%), Gaps = 3/63 (4%)
Frame = +2
Query: 159 PVLFGERQPTVSKKDVVFTSI---NKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTV 215
P FG KK + + K +Y K + KA K +LL+GPPGTGK+T +
Sbjct: 662 PARFGTLAMEPEKKQEILNDLLKFKKGKEYYAK--VGKAW--KRGYLLYGPPGTGKSTMI 829
Query: 216 VEI 218
I
Sbjct: 830 SAI 838
>TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Arabidopsis
thaliana}, partial (23%)
Length = 1254
Score = 32.0 bits (71), Expect = 0.74
Identities = 32/109 (29%), Positives = 53/109 (48%), Gaps = 6/109 (5%)
Frame = +3
Query: 202 LLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVR--IGHP 259
LL GPPGTGKT + + G+ + NI++ +I + K V+
Sbjct: 81 LLFGPPGTGKTMLAKAV---ATEAGANFI-----NISMSSITSKWFGEGEKYVKAVFSLA 236
Query: 260 ARLLPQVV-DSALDAQVLRGDNSGLANDIRKEMK---VLNGKLLKTKEK 304
+++ P V+ +D+ + R +N G +RK MK ++N L+TKEK
Sbjct: 237 SKIAPSVIFVDEVDSMLGRRENPGEHEAMRK-MKNEFMVNWDGLRTKEK 380
>TC77261 homologue to GP|8777330|dbj|BAA96920.1 26S proteasome AAA-ATPase
subunit RPT3 {Arabidopsis thaliana}, partial (94%)
Length = 1575
Score = 32.0 bits (71), Expect = 0.74
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 11/98 (11%)
Frame = +2
Query: 126 LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDY 185
L K + V HR +AL+ V A I +L +P V+ D+ I K
Sbjct: 449 LLKPSASVALHRHSNALVD----VLPPEADSSISLLSQSEKPDVTYNDIGGCDIQKQ--- 607
Query: 186 SQKDAISKALSSKNVF-----------LLHGPPGTGKT 212
++A+ L+ ++ LL+GPPGTGKT
Sbjct: 608 EIREAVELPLTHHELYKQIGIDPPRGVLLYGPPGTGKT 721
>TC93594 weakly similar to GP|20466452|gb|AAM20543.1 unknown protein
{Arabidopsis thaliana}, partial (40%)
Length = 754
Score = 32.0 bits (71), Expect = 0.74
Identities = 35/123 (28%), Positives = 53/123 (42%), Gaps = 10/123 (8%)
Frame = +1
Query: 198 KNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRIG 257
K +LL+GPPGTGK++ V I + L +I + N V+ R L+ I
Sbjct: 94 KRGYLLYGPPGTGKSSLVAAI--------ANFLKYDIYDIELTN-VKNNAELRKLLIGIT 246
Query: 258 HPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMK----------VLNGKLLKTKEKNTR 307
+ ++ + +D +LD R +S ND KE K L G KEKN
Sbjct: 247 SKSVVVIEDIDCSLDLTGQRKTDS--ENDKEKEEKNEEVNQVAAASLQGLQAADKEKNKA 420
Query: 308 REI 310
++
Sbjct: 421 SQV 429
>TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase [imported]
- Arabidopsis thaliana, partial (74%)
Length = 1193
Score = 31.6 bits (70), Expect = 0.97
Identities = 26/95 (27%), Positives = 46/95 (48%), Gaps = 2/95 (2%)
Frame = +3
Query: 201 FLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVD--NIVERLVPHRVKLVRIGH 258
FLL+GPPGTGK+ + + S + ++S++ E+LV + ++ R
Sbjct: 168 FLLYGPPGTGKSYLAKAV---ATEADSTFFSISSSDLVSKWMGESEKLVSNLFQMARESA 338
Query: 259 PARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKV 293
P+ + +DS L Q G+ S + I+ E+ V
Sbjct: 339 PSIIFVDEIDS-LCGQRGEGNESEASRRIKTELLV 440
>TC88082 similar to GP|10798790|dbj|BAB16439. replication factor C 36kDa
subunit {Oryza sativa (japonica cultivar-group)},
partial (88%)
Length = 1354
Score = 31.2 bits (69), Expect = 1.3
Identities = 11/17 (64%), Positives = 16/17 (93%)
Frame = +1
Query: 202 LLHGPPGTGKTTTVVEI 218
LL+GPPGTGKT+T++ +
Sbjct: 271 LLYGPPGTGKTSTILAV 321
>TC81176 similar to GP|17065122|gb|AAL32715.1 Similar replication factor C
37-kDa subunit {Arabidopsis thaliana}, partial (79%)
Length = 875
Score = 30.8 bits (68), Expect = 1.7
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = +2
Query: 202 LLHGPPGTGKTTTVVEIILQ 221
L +GPPGTGKTTT + I Q
Sbjct: 158 LFYGPPGTGKTTTALAIAHQ 217
>TC90038 similar to PIR|A84451|A84451 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, partial (43%)
Length = 1000
Score = 30.4 bits (67), Expect = 2.2
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = +1
Query: 202 LLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVER 245
LL+GPPGTGK T++V I++E I++ N A+ ER
Sbjct: 289 LLYGPPGTGK-TSLVRAIVEECGANLTIISPNTVNSALAGESER 417
>TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, complete
Length = 1793
Score = 30.4 bits (67), Expect = 2.2
Identities = 37/166 (22%), Positives = 71/166 (42%), Gaps = 9/166 (5%)
Frame = +2
Query: 201 FLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVD--NIVERLVPHRVKLVRIGH 258
FLL+GPPGTGK+ + + S + ++S++ E+LV + ++ R
Sbjct: 686 FLLYGPPGTGKSYLAKAV---ATEADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESA 856
Query: 259 PARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVL-------NGKLLKTKEKNTRREIQ 311
P+ + +DS + G+ S + I+ E+ V + K+L NT +
Sbjct: 857 PSIIFVDEIDSLCGTRG-EGNESEASRRIKTELLVQMQGVGHNDQKVLVLAATNTPYALD 1033
Query: 312 KELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFD 357
+ +R R KR + + D +K + +G + L + F+
Sbjct: 1034QAIR---RRFDKRIYIPLPD-LKARQHMFKVHLGDTPHNLTESDFE 1159
>TC84649 similar to PIR|A84451|A84451 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, partial (12%)
Length = 486
Score = 30.4 bits (67), Expect = 2.2
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = +1
Query: 202 LLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVER 245
LL+GPPGTGK T++V I++E I++ N A+ ER
Sbjct: 271 LLYGPPGTGK-TSLVRAIVEECGANLTIISPNTVNSALAGESER 399
>AW774182 homologue to PIR|F84674|F84 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, partial (27%)
Length = 487
Score = 30.0 bits (66), Expect = 2.8
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 2/71 (2%)
Frame = -1
Query: 201 FLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVD--NIVERLVPHRVKLVRIGH 258
FLL+GPPGTGK+ + K S + ++S++ E+LV + ++ R
Sbjct: 244 FLLYGPPGTGKSYLAKAV---ATKADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESA 74
Query: 259 PARLLPQVVDS 269
P+ + +DS
Sbjct: 73 PSIIFVDEIDS 41
>TC79768 similar to PIR|B96657|B96657 probable replication factor F16M19.6
[imported] - Arabidopsis thaliana, complete
Length = 1246
Score = 29.6 bits (65), Expect = 3.7
Identities = 28/118 (23%), Positives = 56/118 (46%), Gaps = 3/118 (2%)
Frame = +1
Query: 202 LLHGPPGTGKTTTVVEI---ILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRIGH 258
+L GPPGTGKTT+++ + +L R + + A+ + +D + ++ K V +
Sbjct: 235 ILSGPPGTGKTTSILALAHELLGPNYREAVLELNASDDRGIDVVRNKIKMFAQKKVTL-P 411
Query: 259 PARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRT 316
P R ++D A D+ SG +R+ M++ + NT +I + +++
Sbjct: 412 PGRHKVVILDEA-DSM-----TSGAQQALRRTMEIYSNSTRFALACNTSSKIIEPIQS 567
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,918,079
Number of Sequences: 36976
Number of extensions: 168598
Number of successful extensions: 1008
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 998
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1006
length of query: 633
length of database: 9,014,727
effective HSP length: 102
effective length of query: 531
effective length of database: 5,243,175
effective search space: 2784125925
effective search space used: 2784125925
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC143341.10


