

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.8 - phase: 0
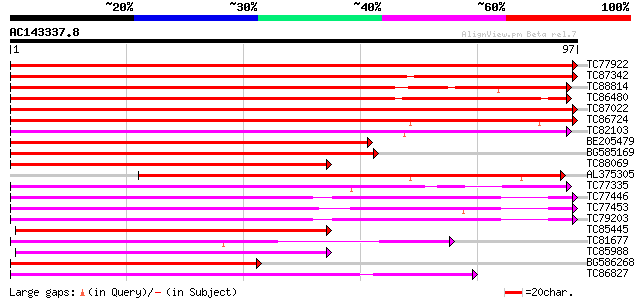
(97 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77922 homologue to PIR|T06448|T06448 GTP-binding protein - gar... 189 1e-49
TC87342 PIR|S41431|S41431 GTP-binding protein ras-like - fava b... 144 4e-36
TC88814 homologue to PIR|T06447|T06447 GTP-binding protein - gar... 117 6e-28
TC86480 homologue to SP|Q40193|R11C_LOTJA Ras-related protein Ra... 103 8e-24
TC87022 homologue to SP|O04486|RB1C_ARATH Ras-related protein Ra... 93 2e-20
TC86724 homologue to PIR|T06445|T06445 GTP-binding protein - gar... 87 1e-18
TC82103 similar to GP|1370144|emb|CAA98178.1 RAB11B {Lotus japon... 74 7e-15
BE205479 homologue to PIR|T06443|T06 GTP-binding protein - garde... 69 2e-13
BG585169 similar to GP|1370152|emb RAB11F {Lotus japonicus}, par... 67 9e-13
TC88069 homologue to GP|1370154|emb|CAA98183.1 RAB11G {Lotus jap... 65 4e-12
AL375305 homologue to PIR|T07059|T070 GTP-binding protein sra1 -... 61 6e-11
TC77335 homologue to GP|1370176|emb|CAA98165.1 RAB2A {Lotus japo... 60 1e-10
TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding prote... 57 1e-09
TC77453 homologue to GP|303732|dbj|BAA02117.1| GTP-binding prote... 53 2e-08
TC79203 homologue to PIR|S41430|S41430 GTP-binding protein ras-... 51 6e-08
TC85445 homologue to GP|1370180|emb|CAA98167.1 RAB5B {Lotus japo... 50 1e-07
TC81677 homologue to PIR|T06446|T06446 GTP-binding protein - gar... 49 3e-07
TC85988 homologue to PIR|S49225|S49225 guanine nucleotide regula... 49 4e-07
BG586268 homologue to PIR|S71585|S71 GTP-binding protein GB2 - A... 47 9e-07
TC86827 homologue to GP|303748|dbj|BAA02115.1| GTP-binding prote... 47 1e-06
>TC77922 homologue to PIR|T06448|T06448 GTP-binding protein - garden pea,
complete
Length = 1182
Score = 189 bits (481), Expect = 1e-49
Identities = 97/97 (100%), Positives = 97/97 (100%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK
Sbjct: 527 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 706
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN
Sbjct: 707 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 817
>TC87342 PIR|S41431|S41431 GTP-binding protein ras-like - fava bean,
complete
Length = 995
Score = 144 bits (364), Expect = 4e-36
Identities = 73/97 (75%), Positives = 85/97 (87%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHLVAV TEDGKS+AE+ESLYFMETSALEATNVENAF EVL+QIYRIVSKK
Sbjct: 438 MLVGNKADLRHLVAVSTEDGKSYAEKESLYFMETSALEATNVENAFAEVLTQIYRIVSKK 617
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
AVE E G ++SVP+ G+ I++K D S LKR GCCS+
Sbjct: 618 AVEGAENG-NASVPAKGEKIDLKNDVSALKRVGCCSS 725
>TC88814 homologue to PIR|T06447|T06447 GTP-binding protein - garden pea,
complete
Length = 1183
Score = 117 bits (294), Expect = 6e-28
Identities = 62/98 (63%), Positives = 80/98 (81%), Gaps = 2/98 (2%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHL AV TED +FAERE+ +FMETSALE+ NVENAFTEVL+QIYR+VSKK
Sbjct: 648 MLVGNKADLRHLRAVSTEDSTAFAERENTFFMETSALESMNVENAFTEVLTQIYRVVSKK 827
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCS 96
A+E G+ +++P GQTI+V ++D S +K+ GCCS
Sbjct: 828 ALEIGD--DPTALPK-GQTIDVGSRDDVSAVKKSGCCS 932
>TC86480 homologue to SP|Q40193|R11C_LOTJA Ras-related protein Rab11C.
{Lotus japonicus}, complete
Length = 1173
Score = 103 bits (258), Expect = 8e-24
Identities = 55/96 (57%), Positives = 71/96 (73%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
M+ GNKSDL HL AV +DG++ AE+E L F+ETSALEATN+E AF +L++IY IVSKK
Sbjct: 555 MMAGNKSDLNHLRAVSEDDGQALAEKEGLSFLETSALEATNIEKAFQTILTEIYHIVSKK 734
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
A+ A E + +S+P G TINV + S+ KR GCCS
Sbjct: 735 ALAAQE-AAGTSLPGQGTTINVADSSANTKR-GCCS 836
>TC87022 homologue to SP|O04486|RB1C_ARATH Ras-related protein Rab11C.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (81%)
Length = 1014
Score = 92.8 bits (229), Expect = 2e-20
Identities = 47/97 (48%), Positives = 66/97 (67%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK+DL+HL AV TED +S+AE+E L F+ETSALEATNVE AF L +IYRI+SKK
Sbjct: 413 MLIGNKTDLKHLRAVATEDAQSYAEKEGLSFIETSALEATNVEKAFQTTLGEIYRIISKK 592
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
++ ++++ G+TI + + CC++
Sbjct: 593 SLSTANEPAAAANIKEGKTIAIGGSETTNTNKPCCTS 703
>TC86724 homologue to PIR|T06445|T06445 GTP-binding protein - garden pea,
complete
Length = 1037
Score = 86.7 bits (213), Expect = 1e-18
Identities = 48/100 (48%), Positives = 67/100 (67%), Gaps = 3/100 (3%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNKSDL + VPTED K FAE+E L+F+ETSAL+ATNVE +F VL++IY IV+KK
Sbjct: 549 ILVGNKSDLENQRDVPTEDAKEFAEKEGLFFLETSALQATNVEASFMTVLTEIYNIVNKK 728
Query: 61 AVEAGEG-GSSSSVPSVGQTINVKEDSSVL--KRFGCCSN 97
+ A E G+ +S +GQ I + + + K CC +
Sbjct: 729 NLAADESQGNGNSASLLGQKIIIPGPAQEIPAKSNMCCQS 848
>TC82103 similar to GP|1370144|emb|CAA98178.1 RAB11B {Lotus japonicus},
partial (80%)
Length = 1513
Score = 74.3 bits (181), Expect = 7e-15
Identities = 42/97 (43%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Frame = +1
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK DL L VPTED FAE + L+F ETSAL NVE AF ++L +I +VSKK
Sbjct: 541 MLVGNKGDLVDLRMVPTEDAVEFAEDQGLFFSETSALTGVNVEGAFMKLLEEINSVVSKK 720
Query: 61 AVEAGE-GGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
A+E G G + + + + S +K+ CS
Sbjct: 721 ALECGNVKGIALKGSKIDVISGAELEISQIKKLSSCS 831
>BE205479 homologue to PIR|T06443|T06 GTP-binding protein - garden pea,
partial (77%)
Length = 499
Score = 69.3 bits (168), Expect = 2e-13
Identities = 32/62 (51%), Positives = 46/62 (73%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK DL ++ V E+GK+ AE E L+FMETSAL++TNV+ AF V+ +IY +S+K
Sbjct: 311 MLVGNKCDLENIREVSIEEGKALAEEEGLFFMETSALDSTNVQTAFEIVIREIYNNISRK 490
Query: 61 AV 62
+
Sbjct: 491 VL 496
>BG585169 similar to GP|1370152|emb RAB11F {Lotus japonicus}, partial (83%)
Length = 668
Score = 67.4 bits (163), Expect = 9e-13
Identities = 33/63 (52%), Positives = 47/63 (74%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+ NKSDL V E+GK FAE+E L FMETSAL+ NVE+AF E++++I+ I+S K
Sbjct: 480 ILVANKSDLSQSREVEKEEGKGFAEKEGLCFMETSALQNLNVEDAFLEMITKIHDIISHK 659
Query: 61 AVE 63
++E
Sbjct: 660 SLE 668
>TC88069 homologue to GP|1370154|emb|CAA98183.1 RAB11G {Lotus japonicus},
partial (79%)
Length = 606
Score = 65.1 bits (157), Expect = 4e-12
Identities = 31/55 (56%), Positives = 42/55 (76%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYR 55
ML+GNK DL ++ V E+GKS AE E L+FMETSAL++TNV+ AF V+ +IY+
Sbjct: 434 MLVGNKCDLDNIRDVSIEEGKSLAESEGLFFMETSALDSTNVKTAFEMVIREIYK 598
>AL375305 homologue to PIR|T07059|T070 GTP-binding protein sra1 - soybean
(fragment), partial (31%)
Length = 509
Score = 61.2 bits (147), Expect = 6e-11
Identities = 35/75 (46%), Positives = 50/75 (66%), Gaps = 2/75 (2%)
Frame = +1
Query: 23 FAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKKAVEAGEG-GSSSSVPSVGQTIN 81
FAE+E L+F+ETSAL+ATNVE AF VL++I+ IV+KK + A E G+ +S G+ I
Sbjct: 1 FAEKEGLFFLETSALQATNVETAFMTVLTEIFNIVNKKNLAADENQGNGNSASLSGKKII 180
Query: 82 VKEDS-SVLKRFGCC 95
V + + K+ CC
Sbjct: 181VPGPAQEIPKKSMCC 225
>TC77335 homologue to GP|1370176|emb|CAA98165.1 RAB2A {Lotus japonicus},
complete
Length = 1121
Score = 60.5 bits (145), Expect = 1e-10
Identities = 41/105 (39%), Positives = 50/105 (47%), Gaps = 9/105 (8%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIV--- 57
MLIGNK DL H AV TE+G+ FA+ L FME SA A NVE AF + IY+ +
Sbjct: 573 MLIGNKCDLAHRRAVSTEEGEQFAKENGLIFMEASAKTAQNVEEAFIKTAGTIYKKIQDG 752
Query: 58 ------SKKAVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
++ G GG PS G+ D GCCS
Sbjct: 753 VFDVSNESYGIKVGYGGIPG--PSGGR------DGPSAAGGGCCS 863
>TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding protein {Pisum
sativum}, complete
Length = 1080
Score = 57.0 bits (136), Expect = 1e-09
Identities = 35/97 (36%), Positives = 52/97 (53%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNKSDL V +E K+FA+ + FMETSA A+NVE AF + ++ I ++
Sbjct: 465 LLVGNKSDLSDKKVVSSETAKAFADEIGIPFMETSAKNASNVEQAFMAMAAE---IKNRM 635
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
A + ++V GQ +N K GCCS+
Sbjct: 636 ASQPANSARPATVQIRGQPVNQKA--------GCCSS 722
>TC77453 homologue to GP|303732|dbj|BAA02117.1| GTP-binding protein {Pisum
sativum}, complete
Length = 944
Score = 52.8 bits (125), Expect = 2e-08
Identities = 36/100 (36%), Positives = 53/100 (53%), Gaps = 3/100 (3%)
Frame = +1
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNKSDL AV + K FA++ + FMETSA +ATNVE AF + + I K
Sbjct: 472 LLVGNKSDLTANRAVSYDTAKEFADQIGIPFMETSAKDATNVEGAFMAMAAAI-----KD 636
Query: 61 AVEAGEGGSSSSVPSV---GQTINVKEDSSVLKRFGCCSN 97
+ + +++ P+V GQ + K GCCS+
Sbjct: 637 RMASQPSANNARPPTVQIRGQPVGQKS--------GCCSS 732
>TC79203 homologue to PIR|S41430|S41430 GTP-binding protein ras-like (clone
vfa-ypt1) - fava bean, complete
Length = 920
Score = 51.2 bits (121), Expect = 6e-08
Identities = 34/97 (35%), Positives = 48/97 (49%)
Frame = +1
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNK DL AV + K+FA+ + FMETSA ++TNVE AF + S I +
Sbjct: 556 LLVGNKCDLTSERAVSYDTAKAFADEIGIPFMETSAKDSTNVEQAFMAMASS---IKDRM 726
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
A + +V GQ + K GCCS+
Sbjct: 727 ASQPTNNARPPTVQIRGQPVGQKS--------GCCSS 813
>TC85445 homologue to GP|1370180|emb|CAA98167.1 RAB5B {Lotus japonicus},
partial (98%)
Length = 1147
Score = 50.1 bits (118), Expect = 1e-07
Identities = 22/54 (40%), Positives = 34/54 (62%)
Frame = +1
Query: 2 LIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYR 55
L+GNK+DL+ V EDG +AE+ ++F+ETSA A N+ F E+ ++ R
Sbjct: 613 LVGNKADLQEKREVAVEDGMDYAEKNGMFFIETSAKTADNINELFEEIAKRLPR 774
>TC81677 homologue to PIR|T06446|T06446 GTP-binding protein - garden pea,
partial (68%)
Length = 667
Score = 48.9 bits (115), Expect = 3e-07
Identities = 31/80 (38%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Frame = +1
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETS----ALEATNVENAFTEVLSQIYRI 56
ML+GNK+DL+HL AV TE+ +FAE+E++YFM V+NAF
Sbjct: 442 MLVGNKADLKHLQAVSTEEATTFAEKENIYFMGRLRWH*EXHVE*VDNAF---------- 591
Query: 57 VSKKAVEAGEGGSSSSVPSV 76
EGGS+SS+ S+
Sbjct: 592 -------RSEGGSTSSIISM 630
>TC85988 homologue to PIR|S49225|S49225 guanine nucleotide regulatory
protein - fava bean, complete
Length = 955
Score = 48.5 bits (114), Expect = 4e-07
Identities = 24/54 (44%), Positives = 32/54 (58%)
Frame = +1
Query: 2 LIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYR 55
L GNKSDL V E+ + +AE L+FMETSA A NV + F E+ ++ R
Sbjct: 487 LAGNKSDLEDKRKVTAEEARVYAEENGLFFMETSAKSAANVNDVFYEIAKRLPR 648
>BG586268 homologue to PIR|S71585|S71 GTP-binding protein GB2 - Arabidopsis
thaliana, partial (49%)
Length = 565
Score = 47.4 bits (111), Expect = 9e-07
Identities = 24/43 (55%), Positives = 28/43 (64%)
Frame = +1
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVE 43
MLIGNK DL H AV E+G+ FA+ L F+E SA A NVE
Sbjct: 124 MLIGNKCDLSHKRAVSKEEGQQFAKEHGLLFLEASARTAQNVE 252
>TC86827 homologue to GP|303748|dbj|BAA02115.1| GTP-binding protein {Pisum
sativum}, complete
Length = 917
Score = 47.0 bits (110), Expect = 1e-06
Identities = 28/80 (35%), Positives = 43/80 (53%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNK DL V T K+FA+ + F+ETSA ++ NVE AF + ++I + +
Sbjct: 473 LLVGNKCDLTDNKLVHTHTAKAFADELGIPFLETSAKDSINVEQAFLTMAAEIKNKMGSQ 652
Query: 61 AVEAGEGGSSSSVPSVGQTI 80
G ++ SV GQ I
Sbjct: 653 --PTGSKSAAESVQMKGQPI 706
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.127 0.336
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,296,949
Number of Sequences: 36976
Number of extensions: 17975
Number of successful extensions: 112
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 110
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 111
length of query: 97
length of database: 9,014,727
effective HSP length: 73
effective length of query: 24
effective length of database: 6,315,479
effective search space: 151571496
effective search space used: 151571496
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 50 (23.9 bits)
Medicago: description of AC143337.8


