

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.12 - phase: 0
(655 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
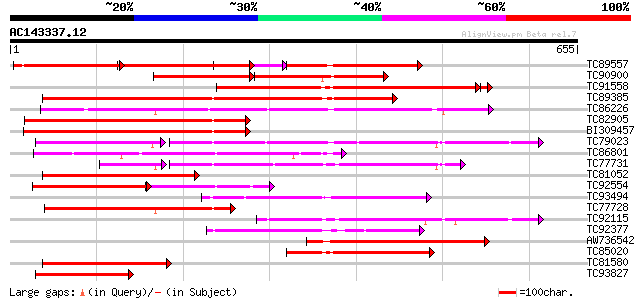
Score E
Sequences producing significant alignments: (bits) Value
TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 210 e-136
TC90900 weakly similar to PIR|D96753|D96753 Similar to disease r... 251 e-118
TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll... 355 6e-98
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 341 5e-94
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 280 1e-75
TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanu... 241 8e-64
BI309457 weakly similar to GP|15787901|gb resistance gene analog... 236 3e-62
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 212 3e-55
TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - ... 210 1e-54
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 201 1e-51
TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 190 2e-48
TC92554 similar to GP|9758876|dbj|BAB09430.1 disease resistance ... 122 1e-44
TC93494 similar to PIR|A54810|A54810 TMV resistance protein N - ... 169 3e-42
TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance... 167 2e-41
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 166 3e-41
TC92377 similar to GP|19774149|gb|AAL99051.1 NBS-LRR-Toll resist... 161 8e-40
AW736542 similar to PIR|T07656|T076 probable resistance protein ... 160 1e-39
TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 159 2e-39
TC81580 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 159 4e-39
TC93827 weakly similar to GP|15787895|gb|AAL07539.1 resistance g... 157 2e-38
>TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein
N {Arabidopsis thaliana}, partial (8%)
Length = 1567
Score = 210 bits (535), Expect(3) = e-136
Identities = 102/159 (64%), Positives = 119/159 (74%)
Frame = +1
Query: 125 KQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLAKLVGWDVR 184
KQ VFP+FYD DPSHV+KQ+GVY+NAFVL FKH+ KV RW M L GWDVR
Sbjct: 523 KQVVFPVFYDVDPSHVKKQNGVYENAFVLHTEAFKHNSGKVARWRTTMTYLGGTAGWDVR 702
Query: 185 NKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWG 244
+PEF I+ IV+ VI +G F G DDLIGIQP +E LE+LLKL SKD R +GIWG
Sbjct: 703 VEPEFEMIEKIVEAVIKKLGRTFSGSTDDLIGIQPHIEALENLLKLSSKDDGCRVLGIWG 882
Query: 245 MAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGGA 283
M GI KTTLA+VLYD++S+QFDA CFIENVSKIY+DGGA
Sbjct: 883 MDGIGKTTLATVLYDKISFQFDACCFIENVSKIYEDGGA 999
Score = 158 bits (399), Expect(3) = e-136
Identities = 81/158 (51%), Positives = 108/158 (68%)
Frame = +3
Query: 320 KFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPL 379
K L+VLDN + LEQ+E+L I P+ L S+III TRD HIL + + V+E L
Sbjct: 1110 KLLIVLDNVEQLEQLEKLDIEPKFLHPRSKIIIITRDKHILQAYGADE------VFEAKL 1271
Query: 380 LNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDA 439
+N DA KL RKAFKS+ P+SG +L P+VL YAQ LPLAV+V+GSFL +R+AN+W
Sbjct: 1272 MNDEDAHKLLCRKAFKSDYPSSGFAELIPKVLVYAQRLPLAVKVLGSFLFSRNANEWSST 1451
Query: 440 LYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFF 477
L + NP N +M LQVS+EGL +++E+FLH+A FF
Sbjct: 1452 LDKFEKNPPNKIMKALQVSYEGLEKDEKEVFLHVAXFF 1565
Score = 158 bits (399), Expect(3) = e-136
Identities = 83/128 (64%), Positives = 98/128 (75%)
Frame = +3
Query: 5 LFRPQSRLTSSDLIHLGQGDIGSSSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTT 64
LF P +++ L G SS+DSNS QSYRYDVFISFRG+DTR++FVDHL+AHL
Sbjct: 168 LFGPMNKMFG--LPREGGTASSSSTDSNSNQSYRYDVFISFRGSDTRNSFVDHLYAHLNR 341
Query: 65 KGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTEL 124
KGIF FKDDK+L+KGE++SPQLLQAIQ SRISIVVFSK+YA ST CL+EMA I E L
Sbjct: 342 KGIFTFKDDKQLQKGEAISPQLLQAIQQSRISIVVFSKDYASSTWCLDEMAAINESRINL 521
Query: 125 KQTVFPIF 132
K + FP F
Sbjct: 522 KTSCFPCF 545
Score = 45.4 bits (106), Expect = 7e-05
Identities = 28/85 (32%), Positives = 43/85 (49%)
Frame = +2
Query: 236 EFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGGATAVQKQILRQTI 295
EF G+W G + L + + ++ ++K + AVQKQI+ QTI
Sbjct: 863 EFWEYGVWMA*G--RQLLPPFCMTKSHFNLMLVVLLKTLAKFMRMVVPVAVQKQIICQTI 1036
Query: 296 DEKNLETYSPSEISGIIRKRLCNKK 320
+EKN++T S +IS +R RLC K
Sbjct: 1037EEKNIDTCSARKISQTMRNRLCKTK 1111
>TC90900 weakly similar to PIR|D96753|D96753 Similar to disease resistance
proteins [imported] - Arabidopsis thaliana, partial (7%)
Length = 826
Score = 251 bits (641), Expect(2) = e-118
Identities = 127/160 (79%), Positives = 143/160 (89%), Gaps = 5/160 (3%)
Frame = +3
Query: 283 ATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPE 342
A ++QKQILRQTIDEK LETYSPSEISGI+RKRLCN+KFLVVLDN DLLEQ+EELAINPE
Sbjct: 354 AVSLQKQILRQTIDEKYLETYSPSEISGIVRKRLCNRKFLVVLDNVDLLEQVEELAINPE 533
Query: 343 LLGKGSRIIITTRDMHIL-----SLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSE 397
L+GKGSR+IITTR+MHIL LS+SH TCVS YEVPLLN+NDAR+LFYRKAFKS+
Sbjct: 534 LVGKGSRMIITTRNMHILRVYGEQLSLSHGTCVS---YEVPLLNNNDARELFYRKAFKSQ 704
Query: 398 DPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWR 437
DP S C+ LTPEVLKY +GLPLA+RVVGSFLCTR+ANQWR
Sbjct: 705 DPASECLNLTPEVLKYVEGLPLAIRVVGSFLCTRNANQWR 824
Score = 193 bits (491), Expect(2) = e-118
Identities = 95/116 (81%), Positives = 101/116 (86%)
Frame = +2
Query: 167 RWVGAMESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELES 226
RW AM LA+LVGWDVRNKPEFREI+NIVQEVI T+GHKF GFADDLI QPRVEELES
Sbjct: 5 RWTKAMGRLAELVGWDVRNKPEFREIENIVQEVIKTLGHKFSGFADDLIATQPRVEELES 184
Query: 227 LLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGG 282
LLKL S D E R +GIWGMAGI KTTLASVLYDR+S QFDASCFIENVSKIY+DGG
Sbjct: 185 LLKLSSDDDELRVVGIWGMAGIGKTTLASVLYDRISSQFDASCFIENVSKIYRDGG 352
>TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll resistance
gene analog protein {Medicago edgeworthii}, partial
(60%)
Length = 942
Score = 355 bits (911), Expect(2) = 6e-98
Identities = 190/306 (62%), Positives = 227/306 (74%), Gaps = 2/306 (0%)
Frame = +1
Query: 240 IGIWGMAGIRKTTLASVLYDRVS--YQFDASCFIENVSKIYKDGGATAVQKQILRQTIDE 297
+GIW M GI KTTLA+VLYD +S YQFDA CF+E+VSKIY+DGGA AVQK+IL QTI E
Sbjct: 1 VGIWRMDGIGKTTLANVLYDTISHQYQFDACCFVEDVSKIYRDGGAIAVQKRILDQTIKE 180
Query: 298 KNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDM 357
KNLE YSPSEISGII RL K L+VLDN D Q++EL INP L GSRIIITTRD
Sbjct: 181 KNLEGYSPSEISGIISNRLYKLKLLLVLDNVDQSVQLQELHINPISLCAGSRIIITTRDK 360
Query: 358 HILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGL 417
HIL + ++ + VYE LLN NDA +L RKAFKS+ +S +L PEVLKYAQGL
Sbjct: 361 HIL---IEYEADI---VYEAELLNDNDAHELLCRKAFKSDYSSSDYEELIPEVLKYAQGL 522
Query: 418 PLAVRVVGSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFF 477
PLA+RV+GSFL R QWR AL +NNPD+ +M VL+ SFEGL ++EIFLH+ACFF
Sbjct: 523 PLAIRVMGSFLYKRKTAQWRAALEGWQNNPDSGIMKVLRSSFEGLELREKEIFLHVACFF 702
Query: 478 KGEKEDYVKRILDACGLHPHIGIQSLIERSFITIRNNEILMHEMLQELGKKIVRQQFPFQ 537
GE+EDYV+RIL ACGL P+IGI + +E+S ITIRN E MHEMLQELG+ VR+ P +
Sbjct: 703 DGEREDYVRRILHACGLQPNIGIPTSVEKSLITIRNQENHMHEMLQELGETNVRETHPDE 882
Query: 538 PGSWSR 543
P WS+
Sbjct: 883 PRLWSK 900
Score = 21.6 bits (44), Expect(2) = 6e-98
Identities = 7/14 (50%), Positives = 9/14 (64%)
Frame = +3
Query: 544 LWLYDDFYSVMMTE 557
+WLY DF M T+
Sbjct: 900 IWLYSDFCHAMTTQ 941
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 341 bits (875), Expect = 5e-94
Identities = 186/412 (45%), Positives = 262/412 (63%), Gaps = 2/412 (0%)
Frame = +3
Query: 39 YDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIV 98
YDVF++FRG DTR+ F + L A L KGI+AF+DD L KGES+ P+LL+ I+ S++ +
Sbjct: 69 YDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRTIEGSQVFVA 248
Query: 99 VFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKF 158
V S+NYA ST CL+E+ I E + V PIFY DPS V+KQSG+Y + F + +F
Sbjct: 249 VLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYWDDFAKHEQRF 428
Query: 159 KHDPNKVMRWVGAMESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQ 218
K DP+KV RW A+ + + GWD+R+K + E++ IVQ ++N + K + DL+GI
Sbjct: 429 KQDPHKVSRWREALNQVGSIAGWDLRDKQQSVEVEKIVQTILNILKCKSSFVSKDLVGIN 608
Query: 219 PRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVSKIY 278
R E L+ L L+S D R IGIWGM G KTTLA LY ++ ++FDASCFI++VSKI+
Sbjct: 609 SRTEALKHQLLLNSVD-GVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFIDDVSKIF 785
Query: 279 K-DGGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEEL 337
+ G QKQIL QT+ ++ + + + +IR RL +K L++LDN D +EQ+E +
Sbjct: 786 RLHDGPIDAQKQILHQTLGIEHHQICNHYSATDLIRHRLSREKTLLILDNVDQVEQLERI 965
Query: 338 AINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSE 397
++ E LG GSRI+I +RD HIL + V D Y+VPLL+ ++ KLF +KAFK E
Sbjct: 966 GVHREWLGAGSRIVIISRDEHILK-----EYKVDVD-YKVPLLDWTESHKLFRQKAFKLE 1127
Query: 398 D-PTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYRLRNNPD 448
L E+L YA GLPL + V+GSFL R+ +W+ AL RLR P+
Sbjct: 1128KIIMKNYQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRPN 1283
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 280 bits (717), Expect = 1e-75
Identities = 178/537 (33%), Positives = 290/537 (53%), Gaps = 14/537 (2%)
Frame = +3
Query: 36 SYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDD--KRLEKGESLSPQLLQAIQSS 93
++ YDVF+ +G DTR F +L L KGI F DD L++ + ++P + I+ S
Sbjct: 81 AFSYDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDKVTPII---IEES 251
Query: 94 RISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVL 153
RI I +FS NYA S+ CL+ + I + V P+F+ +P+ VR +G Y A
Sbjct: 252 RILIPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVRHHTGRYGKALAE 431
Query: 154 LQNKFKHDPNKVMR---WVGAMESLAKLVGW-DVRNKPEFREIKNIVQEVINTMGHKFLG 209
+N+F++D + R W A+ A L + D + E+ I IV+ + N + + L
Sbjct: 432 HENRFQNDTKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGKIVKYISNKISRQSLH 611
Query: 210 FADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASC 269
A +G+Q RV++++SLL + D +GI+G+ G K+TLA +Y+ V+ QF+ C
Sbjct: 612 VATYPVGLQSRVQQVKSLLD-EGPDDGVHMVGIYGIGGSGKSTLARAIYNFVADQFEGLC 788
Query: 270 FIENVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNAD 329
F+E V + Q+ +L +T+ K ++ SE II++RLC KK L++LD+ D
Sbjct: 789 FLEQVRENSASNSLKRFQEMLLSKTLQLK-IKLADVSEGISIIKERLCRKKILLILDDVD 965
Query: 330 LLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLF 389
++Q+ LA + G GSR+IITTRD H+L+ T Y V LN +A +L
Sbjct: 966 NMKQLNALAGGVDWFGPGSRVIITTRDKHLLACHEIEKT------YAVKGLNVTEALELL 1127
Query: 390 YRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYRLRNNPDN 449
AFK++ S K+ V+ YA GLP+ + +VGS L ++ + ++ L P+
Sbjct: 1128RWMAFKNDKVPSSYEKILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNK 1307
Query: 450 NVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRILDACGLHPHIG------IQSL 503
+ +L+VS++ L E++ +FL IAC FKG K + VK I LH H G ++ L
Sbjct: 1308EIQRILKVSYDSLEEEEQSVFLDIACCFKGCKWEKVKEI-----LHAHYGHCINHHVEVL 1472
Query: 504 IERSFIT--IRNNEILMHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTET 558
+E+ I ++ + +H +++ +GK++VR + PF+PG SRLW D + V+ T
Sbjct: 1473VEKCLIDHFEYDSHVSLHNLIENMGKELVRLESPFEPGKRSRLWFEKDIFEVLEENT 1643
>TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum
tuberosum}, partial (22%)
Length = 832
Score = 241 bits (614), Expect = 8e-64
Identities = 124/261 (47%), Positives = 172/261 (65%)
Frame = +1
Query: 18 IHLGQGDIGSSSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLE 77
I + SS+ S + YDVF++FRG DTR+ F D L L TKGI F+D L+
Sbjct: 25 IQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQ 204
Query: 78 KGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADP 137
KGE + P+L +AI+ S++ + +FSKNYA ST CL+E+ I E + V P+FYD DP
Sbjct: 205 KGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDP 384
Query: 138 SHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLAKLVGWDVRNKPEFREIKNIVQ 197
S VRKQSG+Y AFV + +F+ D KV RW A+E + + GWD+R++P REIK IVQ
Sbjct: 385 SEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQ 564
Query: 198 EVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVL 257
++IN + K+ + DL+GI ++ L++ L L+S D RAIGI GM GI KTTLA+ L
Sbjct: 565 KIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVD-GVRAIGICGMGGIGKTTLATTL 741
Query: 258 YDRVSYQFDASCFIENVSKIY 278
Y ++S+QF ASCFI++V+KIY
Sbjct: 742 YGQISHQFSASCFIDDVTKIY 804
>BI309457 weakly similar to GP|15787901|gb resistance gene analog NBS7
{Helianthus annuus}, partial (45%)
Length = 806
Score = 236 bits (601), Expect = 3e-62
Identities = 123/262 (46%), Positives = 170/262 (63%)
Frame = +3
Query: 17 LIHLGQGDIGSSSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRL 76
LI + SS S + YDVF++FRG DTR+ F D L L TKGI F DD L
Sbjct: 6 LIQMASTSNSSSVLGTSSRRNYYDVFVTFRGEDTRNNFTDFLFDALQTKGIIVFSDDTNL 185
Query: 77 EKGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDAD 136
KGES+ P+LL+AI+ S++ + VFS NYA ST CL+E+ I E + V P+FYD D
Sbjct: 186 PKGESIGPELLRAIEGSQVFVAVFSINYASSTWCLQELEKICECVKGSGKHVLPVFYDVD 365
Query: 137 PSHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLAKLVGWDVRNKPEFREIKNIV 196
PS VRKQSG+Y AF+ + +F+ + KV +W A++ + + GWD+R+KP+ EIK IV
Sbjct: 366 PSDVRKQSGIYGEAFIKHEQRFQQEFQKVSKWRDALKQVGSISGWDLRDKPQAGEIKKIV 545
Query: 197 QEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASV 256
Q ++N + +K F+ DL+GI R++ L++ L LDS D RAIGI GM GI KTTLA
Sbjct: 546 QTILNILKYKSSCFSKDLVGIDSRLDGLQNHLLLDSVD-SVRAIGICGMGGIGKTTLAMA 722
Query: 257 LYDRVSYQFDASCFIENVSKIY 278
LYD++S++F AS FI++V + Y
Sbjct: 723 LYDQISHRFSASWFIDDVKQNY 788
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 212 bits (540), Expect = 3e-55
Identities = 145/438 (33%), Positives = 235/438 (53%), Gaps = 6/438 (1%)
Frame = +3
Query: 185 NKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWG 244
N+ E+ I IV+++ N + + L A +G+Q RV+ ++ L S D E +G++G
Sbjct: 684 NRYEYEFIGKIVEDISNRISREPLDVAKYPVGLQSRVQHVKGHLDEKSDD-EVHMVGLYG 860
Query: 245 MAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGGATAVQKQILRQTIDEKNLETYS 304
GI K+TLA +Y+ ++ QF+ CF+ENV +Q+++L +T+ +++
Sbjct: 861 TGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTV-RLDIKLGG 1037
Query: 305 PSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSV 364
S+ II++RLC KK L++LD+ D L+Q+E LA + G GSR+IITTR+ H+L +
Sbjct: 1038 VSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHG 1217
Query: 365 SHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVV 424
T + V LN+ +A +L AFK P+S + L YA GLPLA+ ++
Sbjct: 1218 IEST------HAVEGLNATEALELLRWMAFKENVPSSH-EDILNRALTYASGLPLAIVII 1376
Query: 425 GSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDY 484
GS L R L P+ + +L+VS++ L E++ +FL IAC FKG K
Sbjct: 1377 GSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCKWPE 1556
Query: 485 VKRILDA----CGLHPHIGIQSLIERSFI--TIRNNEILMHEMLQELGKKIVRQQFPFQP 538
VK IL A C +H H+ + L E+S + ++ + +H++++++GK++VRQ+ P +P
Sbjct: 1557 VKEILHAHYGHCIVH-HVAV--LAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEP 1727
Query: 539 GSWSRLWLYDDFYSVMMTETGTNNINAIILDQKEHISEYPQLRAEALSIMRGLKILILLF 598
G SRLW D V+ TGT I I + S+ A M LK I
Sbjct: 1728 GERSRLWFERDIVHVLKKNTGTRKIKMINMKFPSMESDI-DWNGNAFEKMTNLKTFI-TE 1901
Query: 599 HKNFSGSLTFLSNSLQYL 616
+ + S SL +L +SL+ +
Sbjct: 1902 NGHHSKSLEYLPSSLRVM 1955
Score = 119 bits (299), Expect = 3e-27
Identities = 62/153 (40%), Positives = 91/153 (58%), Gaps = 3/153 (1%)
Frame = +1
Query: 31 SNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAI 90
+ S S+ Y VF+SFRGADTR F +L+ LT KGI+ F DD L++G+ ++P L AI
Sbjct: 70 TQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAI 249
Query: 91 QSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNA 150
+ SRI I VFS+NYA S+ CL+E+ I + V P+F DP+ VR +G Y A
Sbjct: 250 EKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEA 429
Query: 151 FVLLQNKFKHDPN---KVMRWVGAMESLAKLVG 180
+ + KF++D + ++ +W A+ A L G
Sbjct: 430 LAVHKKKFQNDKDNTERLQQWKEALSQAANLSG 528
>TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (7%)
Length = 1268
Score = 210 bits (535), Expect = 1e-54
Identities = 131/370 (35%), Positives = 217/370 (58%), Gaps = 8/370 (2%)
Frame = +2
Query: 28 SSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLL 87
+S S+S +YDVFISFRG DTR+ F HLHA L+ + + D R+EKG+ + P+L
Sbjct: 35 ASSSHSAAQKKYDVFISFRGEDTRAGFTSHLHAALSRTYLHTYID-YRIEKGDEVWPELE 211
Query: 88 QAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQT---VFPIFYDADPSHVRKQS 144
+AI+ S + +VVFS+NYA ST CL E+ + E + + V P+FY DPSHVRKQ+
Sbjct: 212 KAIKQSTLFLVVFSENYASSTWCLNELVELMECRNKNEDDNIGVIPVFYHVDPSHVRKQT 391
Query: 145 GVYQNAFVLLQNKFKHDPNKVMR-WVGAMESLAKLVGWDVRN-KPEFREIKNIVQEVINT 202
G Y +A L ++K ++ +K+M+ W A+ A L G+ + E I++I + ++
Sbjct: 392 GSYGSA--LAKHKQENQDDKMMQNWKNALFQAANLSGFHSSTYRTESNMIEDITRALLGK 565
Query: 203 MGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVS 262
+ H++ + + + SL+K DS + IG+WGM G KTTLA+ ++ R S
Sbjct: 566 LNHQYRDELTCNLILDENYWAVRSLIKFDSTTVQI--IGLWGMGGTGKTTLAAAMFQRFS 739
Query: 263 YQFDASCFIENVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFL 322
++++ +CF+E V+++ K G ++L + + E +L +P I +I++RL + K
Sbjct: 740 FKYEGNCFLERVTEVSKKHGINYTCNKLLSKLLGE-DLRIDTPKVIPAMIKRRLRHMKSF 916
Query: 323 VVLD---NADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPL 379
+VLD N++LL+ + + + LG GS +I+TTRD H+L +S D ++YEV
Sbjct: 917 IVLDDVHNSELLQDL--IGVRGGWLGPGSIVIVTTRDKHVL-ISGGID-----EIYEVKK 1072
Query: 380 LNSNDARKLF 389
+NS ++ +LF
Sbjct: 1073MNSQNSLQLF 1102
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 201 bits (510), Expect = 1e-51
Identities = 126/349 (36%), Positives = 205/349 (58%), Gaps = 7/349 (2%)
Frame = +1
Query: 185 NKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWG 244
N+ E++ I+ IV+++ N + H FL A +G+Q R+E+++ LL + S+D E R +G++G
Sbjct: 493 NRYEYKFIEKIVEDISNNINHVFLNVAKYPVGLQSRIEQVKLLLDMGSED-EVRMVGLFG 669
Query: 245 MAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGGATAVQKQILRQTID-EKNLETY 303
G+ K+TLA +Y+ V+ QF+ CF+ NV + +QK++L + + + LE
Sbjct: 670 TGGMGKSTLAKAVYNFVADQFEGVCFLHNVRENSTHKNLKHLQKKLLSKIVKFDGKLEDV 849
Query: 304 SPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLS 363
S E II++RL KK L++LD+ D LEQ+E LA + G GSR+IITTRD H+L+
Sbjct: 850 S--EGIPIIKERLSRKKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLAC- 1020
Query: 364 VSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRV 423
H ++ V E LN +A +L R AFK++ S ++ V+ YA GLPLA+
Sbjct: 1021--HGITSTHAVEE---LNETEALELLRRMAFKNDKVPSTYEEILNRVVTYASGLPLAIVT 1185
Query: 424 VGSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKED 483
+G L R W+ L N P+ ++ +LQVS++ L +++ +FL IAC FKG K
Sbjct: 1186IGDNLFGRKVEDWKRILDEYENIPNKDIQRILQVSYDALEPKEKSVFLDIACCFKGCKWT 1365
Query: 484 YVKRILDA----CGLHPHIGIQSLIERSFI--TIRNNEILMHEMLQELG 526
VK+IL A C H H+G+ L E+S I + ++ +H++++++G
Sbjct: 1366KVKKILHAHYGHCIEH-HVGV--LAEKSLIGHWEYDTQMTLHDLIEDMG 1503
Score = 52.8 bits (125), Expect = 4e-07
Identities = 26/81 (32%), Positives = 42/81 (51%), Gaps = 3/81 (3%)
Frame = +2
Query: 104 YAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPN 163
YA S+ CL+E+ I + V P+FYD +P+H+R QSG Y + +F+++
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEYLTKHEERFQNNEK 181
Query: 164 KVMR---WVGAMESLAKLVGW 181
+ R W A+ A L G+
Sbjct: 182 NMERLRQWKIALTQAANLSGY 244
>TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein
N {Arabidopsis thaliana}, partial (8%)
Length = 656
Score = 190 bits (482), Expect = 2e-48
Identities = 86/181 (47%), Positives = 121/181 (66%)
Frame = +2
Query: 39 YDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIV 98
YDVF++FRG DTR F+DHL A L KGIFAF+DD L+KGES+ P+L++AI+ S++ I
Sbjct: 110 YDVFVTFRGEDTRFNFIDHLFAALQRKGIFAFRDDANLQKGESIPPELIRAIEGSQVFIA 289
Query: 99 VFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKF 158
V SKNY+ ST CL E+ I + + V P+FYD DPS VR Q G+Y AF + F
Sbjct: 290 VLSKNYSSSTWCLRELVHILDCSQVSGRRVLPVFYDVDPSEVRHQKGIYGEAFSKHEQTF 469
Query: 159 KHDPNKVMRWVGAMESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQ 218
+HD + V W A+ + + GWD+R+KP++ EIK IV+E++N +GH F +L+G+
Sbjct: 470 QHDSHVVQSWREALTQVGNISGWDLRDKPQYAEIKKIVEEILNILGHNFSSLPKELVGMN 649
Query: 219 P 219
P
Sbjct: 650 P 652
>TC92554 similar to GP|9758876|dbj|BAB09430.1 disease resistance protein
{Arabidopsis thaliana}, partial (7%)
Length = 1043
Score = 122 bits (305), Expect(2) = 1e-44
Identities = 59/138 (42%), Positives = 89/138 (63%)
Frame = +1
Query: 27 SSSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQL 86
SS ++ S ++ VF++FRG DTR F HL+A L K I F DD+ +E+G+++SP L
Sbjct: 199 SSFGASHSNSKQHHVFLNFRGEDTRYNFTSHLYAALCGKKIHTFMDDEEIERGDNISPTL 378
Query: 87 LQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGV 146
L AI+SS+I +++FS++YA S+ CL+E+ I E + K V P+FY D SHVR Q G
Sbjct: 379 LSAIESSKICVIIFSQDYASSSWCLDELVKIIECSEKKKLVVIPVFYHIDASHVRHQRGT 558
Query: 147 YQNAFVLLQNKFKHDPNK 164
Y +AF ++ F+ N+
Sbjct: 559 YGDAFAKHEDPFQGQSNQ 612
Score = 76.6 bits (187), Expect(2) = 1e-44
Identities = 47/152 (30%), Positives = 82/152 (53%), Gaps = 2/152 (1%)
Frame = +2
Query: 157 KFKHDPNKVMRWVGAMESLAKLVGW-DVRNKPEFREIKNIVQEVINTMGHKFLGFADD-L 214
+F+ + KV W A+ A L GW +N+ E IK IV+ +++ + + L
Sbjct: 590 RFRDNLTKVHMWRTALHKAANLAGWVSEKNRSEAVLIKEIVEVILDKLKCMSPHVQNKGL 769
Query: 215 IGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENV 274
+GI V +ES+L S D IGIWGM GI KTT+A ++ +VSYQ++ F NV
Sbjct: 770 VGISVHVAHVESMLCSGSADVHI--IGIWGMGGIGKTTIADAVFTKVSYQYERYYFAANV 943
Query: 275 SKIYKDGGATAVQKQILRQTIDEKNLETYSPS 306
+ + G +Q ++L +++++++ +P+
Sbjct: 944 RETW--GNRIKLQNEVLADVLEDQSIKISTPT 1033
>TC93494 similar to PIR|A54810|A54810 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (6%)
Length = 876
Score = 169 bits (428), Expect = 3e-42
Identities = 99/266 (37%), Positives = 153/266 (57%)
Frame = +3
Query: 222 EELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDG 281
+++ES LK+ S +E R +GIWGM GI KTTLA LY ++ QF+ C + NV
Sbjct: 12 DQIESKLKIGS--HEVRVLGIWGMGGIGKTTLARALYAKMYSQFEGCCLL-NVMDESNKY 182
Query: 282 GATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINP 341
G V ++L ++E+N+ + + +R+ KK L+VLD + LEQ+E+L
Sbjct: 183 GLNVVPNKLLSSLLEEENIHPDASYIEAPFSERRIGRKKVLIVLDGVETLEQIEDLIPKI 362
Query: 342 ELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTS 401
+ LG GSR+IITTRD HI S VS ++YE+ LN +D+ +L AFK + P +
Sbjct: 363 DGLGPGSRVIITTRDKHIFS-QVS-----KCEIYELKELNKHDSLQLLSLTAFKEKQPKT 524
Query: 402 GCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEG 461
G L+ V+ Y +G PLA++V+G+ L +R W + L +L+ P+ + +VL+ S+
Sbjct: 525 GYEDLSESVIAYCKGNPLALKVLGANLSSRGQEAWENELKKLQKIPNQKIYNVLKFSYAD 704
Query: 462 LHSEDREIFLHIACFFKGEKEDYVKR 487
L + IFL IAC G+ +D+ KR
Sbjct: 705 LDRCQKAIFLDIACLLSGQGKDFCKR 782
>TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (12%)
Length = 765
Score = 167 bits (422), Expect = 2e-41
Identities = 88/225 (39%), Positives = 140/225 (62%), Gaps = 4/225 (1%)
Frame = +1
Query: 41 VFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIVVF 100
VF+SFRG+DTR+ F +L+ L KGI F DD LE+G+ ++P L++AI+ SRI I +F
Sbjct: 94 VFLSFRGSDTRNKFTGNLYKALVDKGIRTFIDDNDLERGDEITPSLVKAIEESRIFIPIF 273
Query: 101 SKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKH 160
S NYA S+ CL+E+ I + VFP+FYD +P+H+R QSG+Y + +F++
Sbjct: 274 SANYASSSFCLDELVHIIHCYKTKSCLVFPVFYDVEPTHIRNQSGIYGEHLTKHEERFQN 453
Query: 161 DPNKVMR---WVGAMESLAKLVGWDVR-NKPEFREIKNIVQEVINTMGHKFLGFADDLIG 216
+ + R W A+ A L G+ + E++ I+ IV+++ N + H FL A +G
Sbjct: 454 NEKNMERLRQWKIALIQAANLSGYHYSPHGYEYKFIEKIVEDISNNINHVFLNVAKYPVG 633
Query: 217 IQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRV 261
+Q R+EE++ LL + S+D E R +G++G G+ K+TLA +Y+ V
Sbjct: 634 LQSRIEEVKLLLDMGSED-EVRMVGLFGTGGMGKSTLAKAVYNFV 765
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 166 bits (419), Expect = 3e-41
Identities = 113/342 (33%), Positives = 189/342 (55%), Gaps = 11/342 (3%)
Frame = +2
Query: 286 VQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLG 345
+Q+++L + + K ++ SE I++RL KK L++LD+ D +EQ+ LA P+ G
Sbjct: 2 LQEELLLKALQLK-IKLGGVSEGIPYIKERLHTKKTLLILDDVDDMEQLHALAGGPDWFG 178
Query: 346 KGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVK 405
+GSR+IITTRD H+L SH +++V E L +A +L AFK+ S
Sbjct: 179 RGSRVIITTRDKHLLR---SHGIESTHEVEE---LYGTEALELLRWMAFKNNKVPSIYED 340
Query: 406 LTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSE 465
+ + YA GLPL + +VGS L + +W+ L P+ + +L+VS++ L E
Sbjct: 341 VLNRAVSYASGLPLVLEIVGSNLFGKTIEEWKGTLDGYEKIPNKKIHQILKVSYDALEEE 520
Query: 466 DREIFLHIACFFKG---EKEDYVKRILDACGLHPHIGIQSLIERSFITIRN------NEI 516
+ +FL IAC FKG E+ +Y+ R + H+ + L E+S + I + E+
Sbjct: 521 QQSVFLDIACCFKGCGWEEFEYILRAHYGHRITHHLVV--LAEKSLVKITHPHYGSIYEL 694
Query: 517 LMHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTETGTNNINAIILD--QKEHI 574
+H++++E+GK++VRQ+ P +PG SRLW DD +V+ TGT+ I I ++ +E +
Sbjct: 695 TLHDLIKEMGKEVVRQESPKEPGERSRLWCEDDIVNVLKENTGTSKIEMIYMNFPSEEFV 874
Query: 575 SEYPQLRAEALSIMRGLKILILLFHKNFSGSLTFLSNSLQYL 616
+ + +A M LK LI + + +FS L +L +SL+ L
Sbjct: 875 ID---KKGKAFKKMTRLKTLI-IENVHFSKGLKYLPSSLRVL 988
>TC92377 similar to GP|19774149|gb|AAL99051.1 NBS-LRR-Toll resistance gene
analog protein {Medicago sativa}, partial (57%)
Length = 945
Score = 161 bits (407), Expect = 8e-40
Identities = 97/252 (38%), Positives = 145/252 (57%)
Frame = +1
Query: 228 LKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGGATAVQ 287
L L+S D + +GI+GM GI KTTLA+ LYDR+S+QFDA CFI+++SK+Y++ G ++
Sbjct: 232 LLLESTDI-IQVVGIYGMGGIGKTTLATALYDRISHQFDACCFIDDISKVYRNEGRLVLK 408
Query: 288 KQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKG 347
+ + + + + +IR +L + + L+VLDN D +EQ E+LA++ E LG G
Sbjct: 409 SKHYIKLLANSTFRYAIFTTKTNLIRLKLGHIRALLVLDNVDQVEQQEKLAVSSECLGAG 588
Query: 348 SRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLT 407
SRI+IT+RD+HIL Y+V E+ F +KAF+ S +LT
Sbjct: 589 SRIVITSRDVHILK---------EYEVDEIV--------N*FCQKAFQGGKIKSNYEELT 717
Query: 408 PEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSEDR 467
+ K +G LC R+ ++W AL RLR NP+ ++MDVL++SFEGL +
Sbjct: 718 CHI-KLC*WPTTGN*SIGLILCGRNISEWTSALARLRENPNKDIMDVLRLSFEGLEESEN 894
Query: 468 EIFLHIACFFKG 479
IFL I C G
Sbjct: 895 VIFLDIVCLLTG 930
Score = 41.2 bits (95), Expect = 0.001
Identities = 30/114 (26%), Positives = 52/114 (45%), Gaps = 4/114 (3%)
Frame = +3
Query: 192 IKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKT 251
I+ IV+E+IN +G++F DL+GI +EELE + Y +W +
Sbjct: 123 IEKIVEEIINRLGYRFSSLPKDLVGIDSLIEELEKVFTFGIN*YYTGCRNLWNGRNRKDD 302
Query: 252 TLASVLYDRVSYQFDASCFIENVSKIYKDG----GATAVQKQILRQTIDEKNLE 301
+ L +S C + + + YK + QKQ L QT+ +++L+
Sbjct: 303 SSYCSL*QNLS---SI*CML--LHR*YKQSL*K*RSIGAQKQALHQTLSKQHLQ 449
>AW736542 similar to PIR|T07656|T076 probable resistance protein - soybean
(fragment), partial (83%)
Length = 656
Score = 160 bits (406), Expect = 1e-39
Identities = 83/214 (38%), Positives = 136/214 (62%), Gaps = 3/214 (1%)
Frame = +2
Query: 344 LGKGSRIIITTRDMHILSLSVSHDTCVSYD-VYEVPLLNSNDARKLFYRKAFKSEDPTSG 402
+G GS IIITTRD +L + + D +YE LN +++R+LF AFK +P+
Sbjct: 29 IGPGSIIIITTRDARLLDI-------LGVDFIYEAEGLNVHESRRLFNWHAFKEANPSEA 187
Query: 403 CVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEGL 462
+ L+ +V+ Y GLPLA+ V+GS+L R +W+ + +L+ P++ + + L++SF+GL
Sbjct: 188 FLILSGDVVSYCGGLPLALEVLGSYLFNRRKREWQSVISKLQKIPNDQIHEKLKISFDGL 367
Query: 463 HSE-DREIFLHIACFFKGEKEDYVKRILDACGLHPHIGIQSLIERSFITI-RNNEILMHE 520
++ IFL + CFF G+ YV IL+ CGLH IGI+ LIERS + + +NN++ MH
Sbjct: 368 EDHMEKNIFLDVCCFFIGKDRAYVTEILNGCGLHADIGIEVLIERSLLKVEKNNKLGMHA 547
Query: 521 MLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVM 554
+L+++G++IVR+ P +P +RLW ++D V+
Sbjct: 548 LLRDMGREIVRESSPEEPEKRTRLWCFEDVVDVL 649
>TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (55%)
Length = 500
Score = 159 bits (403), Expect = 2e-39
Identities = 86/172 (50%), Positives = 118/172 (68%), Gaps = 1/172 (0%)
Frame = +1
Query: 320 KFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPL 379
K L++ DN D +EQ+E++A++ E LG GSRI+I +RD HIL + V VY+VPL
Sbjct: 1 KALLIFDNVDQVEQLEKIAVHREWLGAGSRIVIISRDEHILK---EYGVDV---VYKVPL 162
Query: 380 LNSNDARKLFYRKAFKSEDPT-SGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRD 438
+NS D+ +LF RKAFK E S L E+L YA+GLPLA++V+GSFL +W+
Sbjct: 163 MNSTDSYELFCRKAFKVEKIIMSDYQNLANEILDYAKGLPLAIKVLGSFLFGHSVAEWKS 342
Query: 439 ALYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRILD 490
AL RLR +P N+VMDVL +SF+GL +++EIFL IACFF + E YVK +L+
Sbjct: 343 ALARLRESPHNDVMDVLHLSFDGLDEKEKEIFLDIACFFNSQPEKYVKNVLN 498
>TC81580 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (84%)
Length = 827
Score = 159 bits (401), Expect = 4e-39
Identities = 75/148 (50%), Positives = 103/148 (68%)
Frame = +2
Query: 39 YDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIV 98
YDVF +FRG DTR+ F D L L TKGIFAF+DD L+KGES+ P+LL+AI+ SR+ +
Sbjct: 113 YDVFGTFRGEDTRNNFTDFLFDALETKGIFAFRDDTNLQKGESIEPELLRAIEGSRVFVA 292
Query: 99 VFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKF 158
VFS+NYA ST CL+E+ I + ++ + P+FYD DPS VRKQSG+Y AFV + +F
Sbjct: 293 VFSRNYASSTWCLQELEKICKCVQRSRKHILPVFYDVDPSVVRKQSGIYCEAFVKHEQRF 472
Query: 159 KHDPNKVMRWVGAMESLAKLVGWDVRNK 186
+ D V RW A++ + + GWD+R+K
Sbjct: 473 QQDFEMVSRWREALKHVGSISGWDLRDK 556
>TC93827 weakly similar to GP|15787895|gb|AAL07539.1 resistance gene analog
NBS4 {Helianthus annuus}, partial (26%)
Length = 685
Score = 157 bits (396), Expect = 2e-38
Identities = 78/114 (68%), Positives = 89/114 (77%)
Frame = +1
Query: 30 DSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQA 89
D YRYDVFISFRG DTR TF DHL+AHLT KGIF F DD++LEKGES+S QL QA
Sbjct: 277 DRTQSYRYRYDVFISFRGIDTRKTFADHLYAHLTRKGIFTFMDDQQLEKGESISLQLRQA 456
Query: 90 IQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQ 143
IQ SR+SIVVFS++YA ST CL+EM IA+ T+LKQ V P+FY DPSHVRKQ
Sbjct: 457 IQQSRLSIVVFSEDYAGSTWCLDEMTVIADCLTKLKQIVLPVFYYVDPSHVRKQ 618
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,027,036
Number of Sequences: 36976
Number of extensions: 272891
Number of successful extensions: 1662
Number of sequences better than 10.0: 195
Number of HSP's better than 10.0 without gapping: 1498
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1520
length of query: 655
length of database: 9,014,727
effective HSP length: 102
effective length of query: 553
effective length of database: 5,243,175
effective search space: 2899475775
effective search space used: 2899475775
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC143337.12


