

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142506.2 + phase: 1 /partial
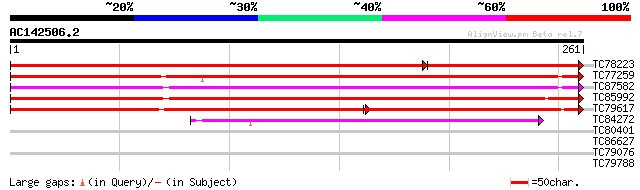
(261 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78223 weakly similar to GP|9758310|dbj|BAB08784.1 porin-like p... 376 e-141
TC77259 similar to PIR|T12558|T12558 porin - common ice plant, p... 213 8e-56
TC87582 similar to PIR|S36454|S36454 porin por1 - garden pea, co... 209 1e-54
TC85992 similar to GP|5031279|gb|AAD38145.1| porin {Prunus armen... 196 8e-51
TC79617 weakly similar to GP|9757871|dbj|BAB08458.1 porin-like p... 137 5e-50
TC84272 weakly similar to PIR|S46959|S46959 porin I 36K - potat... 84 5e-17
TC80401 weakly similar to GP|17385693|dbj|BAB78644. putative Cf2... 29 2.1
TC86627 similar to PIR|T02334|T02334 probable urease accessory p... 28 4.6
TC79076 weakly similar to PIR|E84452|E84452 hypothetical protein... 27 7.9
TC79788 weakly similar to GP|21554189|gb|AAM63268.1 putative leu... 27 7.9
>TC78223 weakly similar to GP|9758310|dbj|BAB08784.1 porin-like protein
{Arabidopsis thaliana}, partial (82%)
Length = 1503
Score = 376 bits (965), Expect(2) = e-141
Identities = 190/190 (100%), Positives = 190/190 (100%)
Frame = +2
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV
Sbjct: 161 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 340
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI
Sbjct: 341 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 520
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG
Sbjct: 521 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 700
Query: 181 LTVAAELAHK 190
LTVAAELAHK
Sbjct: 701 LTVAAELAHK 730
Score = 142 bits (359), Expect(2) = e-141
Identities = 71/71 (100%), Positives = 71/71 (100%)
Frame = +3
Query: 191 LSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSAEYDSKKIIGS 250
LSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSAEYDSKKIIGS
Sbjct: 732 LSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSAEYDSKKIIGS 911
Query: 251 PAKFGLALSLK 261
PAKFGLALSLK
Sbjct: 912 PAKFGLALSLK 944
>TC77259 similar to PIR|T12558|T12558 porin - common ice plant, partial
(98%)
Length = 1113
Score = 213 bits (541), Expect = 8e-56
Identities = 116/263 (44%), Positives = 162/263 (61%), Gaps = 2/263 (0%)
Frame = +1
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDYN D KF+L+ S TG+ LTA+ K+ + F+ D+NT K NVT D+KV+TDSN+
Sbjct: 112 LLYKDYNTDQKFTLTTYSPTGIALTASSTKKGELFLADINTQLKHKNVTTDIKVDTDSNL 291
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDH--KSGKLDVQYLHPHAAIDSSIGLNPAPKLELSA 118
T +T+ + + K LSF IP+ KSGK +VQYLH +A I +SIGL P S
Sbjct: 292 FTTITVTEP--APGLKTILSFRIPEPELKSGKAEVQYLHDYAGISASIGLTANPVANFSG 465
Query: 119 AIGSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRP 178
IG+ +++G ++ +DT + F+ NAG+ F K D A+L L +K +L ASY H V+
Sbjct: 466 VIGNNVVALGGDLSYDTKTGVFTRLNAGLNFTKDDLIASLTLNEKANALNASYYHVVNPV 645
Query: 179 DGLTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITL 238
V AE++H+ S+ EN T GT +ID T K RF++ G A+ Q WRP S T+
Sbjct: 646 TKTAVGAEVSHRFSTKENTLTLGTQHAIDSLTTAKLRFNNFGLASGLIQHEWRPKSFFTI 825
Query: 239 SAEYDSKKIIGSPAKFGLALSLK 261
S E D+K I S A+ G+AL+LK
Sbjct: 826 SGEVDTKAIEKS-ARIGIALALK 891
>TC87582 similar to PIR|S36454|S36454 porin por1 - garden pea, complete
Length = 1307
Score = 209 bits (531), Expect = 1e-54
Identities = 107/261 (40%), Positives = 158/261 (59%)
Frame = +1
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDYN D KF++S S TG+ +T++G K+ + FVGD+NT K+ N T D+KV+T+SN+
Sbjct: 112 LLFKDYNSDQKFTVSTYSPTGVAITSSGTKKGELFVGDVNTQLKNKNYTADIKVDTESNL 291
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+ + + K +S +PD SGK+++QYLH +A I +IGL P + LS
Sbjct: 292 FTTITVTEPA--PGVKAIISCKVPDPASGKVELQYLHDYAGISGNIGLKANPVVNLSGVF 465
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ ++ G ++ +DT + N G++F K D L L +KG L ASY H ++
Sbjct: 466 GTNALAFGGDISYDTKLRELTKSNVGVSFIKDDLLGALTLNEKGDVLNASYYHVINPVTN 645
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V ++ H+ S+ EN FT G ++DP T LK R ++ KA+ Q WRP SLIT+S
Sbjct: 646 TAVGVDMGHRFSTRENNFTLGVQHALDPLTTLKGRITNSAKASALIQHEWRPKSLITIST 825
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GL+L LK
Sbjct: 826 ELDTKAIEKS-AKVGLSLVLK 885
>TC85992 similar to GP|5031279|gb|AAD38145.1| porin {Prunus armeniaca},
partial (88%)
Length = 1293
Score = 196 bits (498), Expect = 8e-51
Identities = 103/261 (39%), Positives = 161/261 (61%)
Frame = +1
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDY DHKF+++ + G+ +T+ G+++ + F+ D++T K+ NVT D+KV+T+SN+
Sbjct: 118 LLFKDYQNDHKFTITTQTLAGVEITSAGVRKGEAFLADVSTKVKNKNVTTDIKVDTNSNL 297
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ K SF P+ KSGK+++QYLH +A I +SIGL P + +SA +
Sbjct: 298 RTTITVDETV--PGLKTIFSFIYPEQKSGKVELQYLHDYAGISTSIGLTATPTVNISAVL 471
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +S+G++V F+T+S S + N G+ D A+L + D+G SL ASY H V
Sbjct: 472 GNNLVSVGSDVSFETSSGSLNKCNFGLNVTHADLIASLTVNDRGDSLNASYYHVVSPLTN 651
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL+H SS+EN T GT ++DP T+LK + ++ GKA+ Q W + +L
Sbjct: 652 TAVGAELSHSFSSNENVLTIGTQHALDPVTLLKAKVNNYGKASALIQHDWSRQARFSLVG 831
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+ I AK GLA +LK
Sbjct: 832 EVDT-AAIQKTAKVGLACALK 891
>TC79617 weakly similar to GP|9757871|dbj|BAB08458.1 porin-like protein
{Arabidopsis thaliana}, partial (97%)
Length = 1208
Score = 137 bits (345), Expect(2) = 5e-50
Identities = 72/164 (43%), Positives = 102/164 (61%)
Frame = +2
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL KDYN D K ++S S+TG+ LT+T LK+ GD+ YK N +DVK+ T S +
Sbjct: 119 LLTKDYNSDQKLTVSSYSTTGVALTSTALKKGGLSTGDVAGQYKYKNTLIDVKIETASII 298
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
+T +T D L S K S +PD+ SGKL+VQY H HA + S L +P ++SA +
Sbjct: 299 NTTLTFTD--LFPSTKTIASIKLPDYSSGKLEVQYFHDHATLTSVATLKQSPIFDVSATV 472
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKG 164
GS ++ GAE G+DT S SF+ YN GI+F K D SA++++ +G
Sbjct: 473 GSPTLAFGAEAGYDTKSGSFTKYNTGISFTKLDSSASVIIWRQG 604
Score = 77.8 bits (190), Expect(2) = 5e-50
Identities = 38/100 (38%), Positives = 61/100 (61%)
Frame = +3
Query: 162 DKGQSLKASYIHYVDRPDGLTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGK 221
DKG S+K SY+H++D+ ++ + S+++N T G S ++DP T +K RF++ GK
Sbjct: 597 DKGDSIKVSYLHHLDQIKKSAAVVDITRRFSTNQNTITVGGSFAVDPLTQIKARFNNQGK 776
Query: 222 AAFQCQRAWRPNSLITLSAEYDSKKIIGSPAKFGLALSLK 261
Q P S++T+S E D+K + P KFGLA++LK
Sbjct: 777 VGGLLQHELFPKSVLTISGEVDTKALEKKP-KFGLAIALK 893
>TC84272 weakly similar to PIR|S46959|S46959 porin I 36K - potato, partial
(7%)
Length = 759
Score = 84.0 bits (206), Expect = 5e-17
Identities = 52/167 (31%), Positives = 81/167 (48%), Gaps = 6/167 (3%)
Frame = +2
Query: 83 IPDHKSGKLDVQYLHPHAAIDSSIGL------NPAPKLELSAAIGSKDISMGAEVGFDTT 136
+PD SGK+++QYL+ I IGL P ++LS +G+ +S+GA V
Sbjct: 242 VPD--SGKVELQYLNRFTGITGCIGLLGNEEGAYDPVVKLSGLLGTSILSLGANVALHIP 415
Query: 137 SASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDGLTVAAELAHKLSSSEN 196
+ S + AG N A+L L D +LKA+ H V+ +A ++ H LS E
Sbjct: 416 TRSITKLTAGFGLNTAFLEASLTLHDSFDTLKATIYHEVNPLTQTAIATQVKHSLSLKET 595
Query: 197 RFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSAEYD 243
T G + P+T+LK RF GK Q+ + +T+++E D
Sbjct: 596 GVTIGAQHAFFPETLLKARFDSSGKVGALIQQGFWQRFFVTMASEID 736
>TC80401 weakly similar to GP|17385693|dbj|BAB78644. putative Cf2/Cf5
disease resistance protein homolog {Oryza sativa
(japonica cultivar-group)}, partial (9%)
Length = 1481
Score = 28.9 bits (63), Expect = 2.1
Identities = 42/149 (28%), Positives = 67/149 (44%), Gaps = 12/149 (8%)
Frame = +3
Query: 70 SLSHSKKVALSFNIPD----HKSGKL-DVQYL-----HPHAAIDSSIG-LNPAPKLELSA 118
SLSH K + LS+N+ D H+ G L ++Q+L + +I S +G L +L LS
Sbjct: 504 SLSHLKYLNLSYNLLDGLIPHRLGDLSNLQFLDLSNNYLEGSIPSQLGKLTNLQELYLSG 683
Query: 119 -AIGSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDR 177
I ++D + G + T+ ++ N ++ L + K +L+ + Y D
Sbjct: 684 LTIDNEDHNGGQWLSNLTSLTHLHMWSIS---NLNKSNSWLKMVGKLPNLRELSLRYCDL 854
Query: 178 PDGLTVAAELAHKLSSSENRFTFGTSQSI 206
D H LS S +F F TS SI
Sbjct: 855 SD------HFIHSLSQS--KFNFSTSLSI 917
>TC86627 similar to PIR|T02334|T02334 probable urease accessory protein
[imported] - Arabidopsis thaliana, partial (93%)
Length = 1246
Score = 27.7 bits (60), Expect = 4.6
Identities = 37/141 (26%), Positives = 56/141 (39%)
Frame = +3
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
L+ +D+N + F++ I G G TA L +L Y VT D+ D
Sbjct: 237 LINRDFN-ERAFTVGIGGPVGTGKTALMLA----LCQNLRDRYSLAAVTNDIFTKEDGEF 401
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
K H +P+ + ++ PHAAI I +N P ELS +
Sbjct: 402 LVK---------HKA-------LPEERIRAVETGGC-PHAAIREDISINLGPLEELS-NL 527
Query: 121 GSKDISMGAEVGFDTTSASFS 141
D+ + E G D +A+FS
Sbjct: 528 YKADLLL-CESGGDNLAANFS 587
>TC79076 weakly similar to PIR|E84452|E84452 hypothetical protein At2g03810
[imported] - Arabidopsis thaliana, partial (6%)
Length = 901
Score = 26.9 bits (58), Expect = 7.9
Identities = 28/114 (24%), Positives = 46/114 (39%)
Frame = +1
Query: 107 GLNPAPKLELSAAIGSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQS 166
GL+P+PKLE ++ +G +V S S+ + G + +M A QS
Sbjct: 529 GLDPSPKLEFYDSMLDLKNGLGTKVTESINPVSRSSNDVG---------SFMMFATDKQS 681
Query: 167 LKASYIHYVDRPDGLTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDG 220
+K S + P L K S + F T +P+ L+ + +DG
Sbjct: 682 VKKSVTSPISNP-------LLVDKFQDSLDVFVDRTEAECEPE--LEICYKEDG 816
>TC79788 weakly similar to GP|21554189|gb|AAM63268.1 putative leucine-rich
repeat disease resistance protein {Arabidopsis
thaliana}, partial (52%)
Length = 1484
Score = 26.9 bits (58), Expect = 7.9
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 1/37 (2%)
Frame = +2
Query: 186 ELAHKLSSSENRFTFGTSQSIDPKTVLK-TRFSDDGK 221
E +HK SS+NRF ++ DPKT + TR + GK
Sbjct: 593 EFSHK--SSQNRFELQ*TRRFDPKTTTQFTRTGNQGK 697
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,775,106
Number of Sequences: 36976
Number of extensions: 80175
Number of successful extensions: 303
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 299
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 300
length of query: 261
length of database: 9,014,727
effective HSP length: 94
effective length of query: 167
effective length of database: 5,538,983
effective search space: 925010161
effective search space used: 925010161
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC142506.2


