

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142498.7 + phase: 0
(1705 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
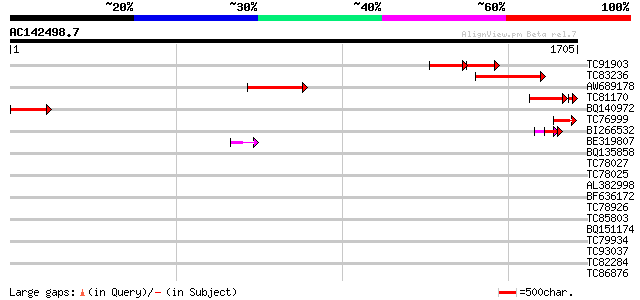
Score E
Sequences producing significant alignments: (bits) Value
TC91903 homologue to PIR|T06779|T06779 clathrin heavy chain - so... 237 e-116
TC83236 homologue to PIR|T06779|T06779 clathrin heavy chain - so... 404 e-112
AW689178 homologue to PIR|T06779|T0 clathrin heavy chain - soybe... 365 e-100
TC81170 homologue to PIR|T06779|T06779 clathrin heavy chain - so... 236 6e-74
BQ140972 similar to PIR|T06779|T0 clathrin heavy chain - soybean... 248 1e-65
TC76999 homologue to GP|19749359|gb|AAD28640.2 geranylgeranyl hy... 116 7e-26
BI266532 weakly similar to GP|12321552|gb clathrin heavy chain ... 62 2e-09
BE319807 similar to GP|12321552|gb clathrin heavy chain putativ... 53 9e-07
BQ135858 similar to GP|23491723|db formin homology protein A {Di... 42 0.002
TC78027 similar to PIR|D86327|D86327 protein F18O14.17 [imported... 37 0.012
TC78025 37 0.013
AL382998 similar to GP|23491723|db formin homology protein A {Di... 38 0.039
BF636172 37 0.086
TC78926 similar to SP|P93231|VP41_LYCES Vacuolar assembly protei... 36 0.11
TC85803 similar to GP|13785471|dbj|BAB43909. phosphoenolpyruvate... 35 0.19
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 35 0.25
TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 prote... 35 0.25
TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV ... 35 0.25
TC82284 weakly similar to GP|11119510|gb|AAF37724.2 diaphanous p... 35 0.25
TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box pro... 35 0.33
>TC91903 homologue to PIR|T06779|T06779 clathrin heavy chain - soybean,
partial (12%)
Length = 640
Score = 237 bits (604), Expect(2) = e-116
Identities = 114/114 (100%), Positives = 114/114 (100%)
Frame = +2
Query: 1262 ANSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESG 1321
ANSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESG
Sbjct: 2 ANSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESG 181
Query: 1322 LGLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKE 1375
LGLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKE
Sbjct: 182 LGLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKE 343
Score = 201 bits (511), Expect(2) = e-116
Identities = 97/100 (97%), Positives = 98/100 (98%)
Frame = +3
Query: 1374 KELTYLYIQYDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDL 1433
+ LTYLYIQYDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDL
Sbjct: 339 RNLTYLYIQYDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDL 518
Query: 1434 INDVLNVLALRVDHARVVDIMRKAGHLRLVKPYMVAVQSN 1473
INDVLNVLALRVDHARVVDIMRK GHLRLVKPYMVAVQSN
Sbjct: 519 INDVLNVLALRVDHARVVDIMRKXGHLRLVKPYMVAVQSN 638
>TC83236 homologue to PIR|T06779|T06779 clathrin heavy chain - soybean,
partial (12%)
Length = 635
Score = 404 bits (1038), Expect = e-112
Identities = 199/211 (94%), Positives = 202/211 (95%)
Frame = +2
Query: 1401 WDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALRVDHARVVDIMRKAGHL 1460
WDHMQFKDV KVANVELYYKAVHFYL+EHPDLIND+LNVLALRVDH RVVDIMRKAGHL
Sbjct: 2 WDHMQFKDVAVKVANVELYYKAVHFYLEEHPDLINDILNVLALRVDHTRVVDIMRKAGHL 181
Query: 1461 RLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELL 1520
RLVKPYMVAVQS+NVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELL
Sbjct: 182 RLVKPYMVAVQSSNVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELL 361
Query: 1521 EMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGERELAEELLVYFIDQGKKEC 1580
EMRRVAAYIYKKAGRWKQ IALSKKDNLYKDAMETASQ GERELAEEL VYFIDQGKKEC
Sbjct: 362 EMRRVAAYIYKKAGRWKQPIALSKKDNLYKDAMETASQYGERELAEELTVYFIDQGKKEC 541
Query: 1581 FASCLFVCYDLIRVDVALELAWMHNMIDFAF 1611
FASCLF CYDLIR DV LELAWMHNMIDFAF
Sbjct: 542 FASCLFGCYDLIRADVVLELAWMHNMIDFAF 634
>AW689178 homologue to PIR|T06779|T0 clathrin heavy chain - soybean, partial
(10%)
Length = 545
Score = 365 bits (936), Expect = e-100
Identities = 180/181 (99%), Positives = 180/181 (99%)
Frame = +3
Query: 715 KIFEQFRSYEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKT 774
KIFEQFRSYEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKT
Sbjct: 3 KIFEQFRSYEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKT 182
Query: 775 KNFLMEAKLPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQL 834
KNFLMEAKLPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQL
Sbjct: 183 KNFLMEAKLPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQL 362
Query: 835 LDDECPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNAL 894
LDDECPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQ VHVHNAL
Sbjct: 363 LDDECPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQXVHVHNAL 542
Query: 895 G 895
G
Sbjct: 543 G 545
>TC81170 homologue to PIR|T06779|T06779 clathrin heavy chain - soybean,
partial (7%)
Length = 718
Score = 236 bits (602), Expect(2) = 6e-74
Identities = 115/115 (100%), Positives = 115/115 (100%)
Frame = +3
Query: 1563 ELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYT 1622
ELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYT
Sbjct: 3 ELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYT 182
Query: 1623 GKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMG 1677
GKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMG
Sbjct: 183 GKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMG 347
Score = 62.0 bits (149), Expect(2) = 6e-74
Identities = 24/25 (96%), Positives = 25/25 (100%)
Frame = +2
Query: 1681 APPPPMGGGMGMPPMPPFGMPMGGY 1705
+PPPPMGGGMGMPPMPPFGMPMGGY
Sbjct: 356 SPPPPMGGGMGMPPMPPFGMPMGGY 430
>BQ140972 similar to PIR|T06779|T0 clathrin heavy chain - soybean, partial
(7%)
Length = 457
Score = 248 bits (633), Expect = 1e-65
Identities = 123/125 (98%), Positives = 123/125 (98%)
Frame = +1
Query: 1 MAAAANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPN 60
MAAAANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPN
Sbjct: 82 MAAAANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPN 261
Query: 61 QPLRRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKW 120
QPLRRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKW
Sbjct: 262 QPLRRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKW 441
Query: 121 ISPKL 125
I P L
Sbjct: 442 IXPXL 456
>TC76999 homologue to GP|19749359|gb|AAD28640.2 geranylgeranyl hydrogenase
{Glycine max}, complete
Length = 2228
Score = 116 bits (291), Expect = 7e-26
Identities = 56/72 (77%), Positives = 62/72 (85%), Gaps = 1/72 (1%)
Frame = +3
Query: 1634 ESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGG-GMGGGYAPPPPMGGGMGM 1692
+ QNEVK+KE+EEK+V+AQQNMYAQLLPLALPAPPMPGMGG G GG Y PPPP+ GMGM
Sbjct: 1671 KKQNEVKSKEKEEKDVVAQQNMYAQLLPLALPAPPMPGMGGPGYGGSYGPPPPV-SGMGM 1847
Query: 1693 PPMPPFGMPMGG 1704
P MPPFGMP G
Sbjct: 1848 PQMPPFGMPPMG 1883
>BI266532 weakly similar to GP|12321552|gb clathrin heavy chain putative
{Arabidopsis thaliana}, partial (5%)
Length = 562
Score = 62.0 bits (149), Expect = 2e-09
Identities = 31/74 (41%), Positives = 43/74 (57%)
Frame = +1
Query: 1577 KKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGKVDELVKHKIESQ 1636
KKECFASCL CY+LIR DVALELAW +NM+DF R L K ++
Sbjct: 244 KKECFASCLSTCYELIRPDVALELAWKNNMMDFPSLISYSLFRNIRAM*MSLKKTELNQN 423
Query: 1637 NEVKAKEQEEKEVI 1650
+ K+++E+ ++
Sbjct: 424 LRRRLKKRKERTLL 465
Score = 59.7 bits (143), Expect = 9e-09
Identities = 31/56 (55%), Positives = 38/56 (67%), Gaps = 2/56 (3%)
Frame = +3
Query: 1609 FAFPYLLQFIREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQ--NMYAQLLPL 1662
F+FPYLLQ I EYT VDEL K ++ES+ E K KE+E K +IAQQ + L PL
Sbjct: 339 FSFPYLLQSIPEYTSNVDELKKDRVESEFEKKVKEKERKNIIAQQVPKSFTSLSPL 506
>BE319807 similar to GP|12321552|gb clathrin heavy chain putative
{Arabidopsis thaliana}, partial (1%)
Length = 386
Score = 53.1 bits (126), Expect = 9e-07
Identities = 33/84 (39%), Positives = 43/84 (50%)
Frame = +3
Query: 665 ALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRSYE 724
A VEF GTLS+E LE MKDL L+NLRGN+QII Q+ K
Sbjct: 69 AFVEFVGTLSEE*VLESMKDLSLLNLRGNIQIIAQMTK---------------------- 182
Query: 725 GLYFFLGSYLSSSEDPDIHFKYIE 748
+ + ++ PDI+FKY+E
Sbjct: 183 -------NCMMKAKLPDIYFKYVE 233
Score = 30.0 bits (66), Expect(3) = 0.001
Identities = 13/19 (68%), Positives = 15/19 (78%)
Frame = +1
Query: 848 ILSVRSLLPVEPLVAECEK 866
IL SL+P+EPLV ECEK
Sbjct: 286 IL*AHSLVPIEPLVKECEK 342
Score = 27.3 bits (59), Expect(3) = 0.001
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = +2
Query: 821 KVNPGNAPLVVGQLLDDE 838
KVN N+ +VVG+LLDDE
Sbjct: 233 KVNRTNSAIVVGKLLDDE 286
Score = 23.5 bits (49), Expect(3) = 0.001
Identities = 9/14 (64%), Positives = 12/14 (85%)
Frame = +3
Query: 772 EKTKNFLMEAKLPD 785
+ TKN +M+AKLPD
Sbjct: 171 QMTKNCMMKAKLPD 212
>BQ135858 similar to GP|23491723|db formin homology protein A {Dictyostelium
discoideum}, partial (4%)
Length = 1033
Score = 42.4 bits (98), Expect = 0.002
Identities = 20/33 (60%), Positives = 21/33 (63%)
Frame = +2
Query: 1672 MGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
+GGG GGG PPP GGG G PP PP P GG
Sbjct: 434 LGGGGGGGSPPPPGGGGGGGTPPPPP---PRGG 523
Score = 41.2 bits (95), Expect = 0.003
Identities = 19/28 (67%), Positives = 19/28 (67%)
Frame = -1
Query: 1670 PGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P GGG GGG PPPP GGG G PP PP
Sbjct: 520 PPRGGG-GGGPPPPPPPGGGGGPPPPPP 440
Score = 39.7 bits (91), Expect = 0.010
Identities = 18/28 (64%), Positives = 18/28 (64%)
Frame = -3
Query: 1670 PGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P GGG GG PPPP GGG G PP PP
Sbjct: 521 PPAGGGGGGSPPPPPPRGGG-GTPPPPP 441
Score = 39.3 bits (90), Expect = 0.013
Identities = 19/36 (52%), Positives = 21/36 (57%)
Frame = +2
Query: 1666 APPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
+PP PG GGG GG PPPP GG + PPF P
Sbjct: 458 SPPPPGGGGG-GGTPPPPPPRGGPLFFFLTPPFSFP 562
Score = 39.3 bits (90), Expect = 0.013
Identities = 24/54 (44%), Positives = 24/54 (44%), Gaps = 14/54 (25%)
Frame = +1
Query: 1665 PAPPMPGMG--------------GGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P PP G G GG GGG PPPP GGG G P PP P GG
Sbjct: 367 PPPPPGGGGNPIGGGGGGGNFFFGGGGGGGVPPPPRGGGGGGDPPPP--PPAGG 522
Score = 38.5 bits (88), Expect = 0.023
Identities = 17/28 (60%), Positives = 17/28 (60%)
Frame = -2
Query: 1670 PGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P GGG GG PPPP GGG PP PP
Sbjct: 522 PPRGGGGGGVPPPPPPPGGGGDPPPPPP 439
Score = 37.4 bits (85), Expect = 0.050
Identities = 23/53 (43%), Positives = 23/53 (43%), Gaps = 18/53 (33%)
Frame = +1
Query: 1667 PPMPGMGG---------------GMGGGYAPPPPMGGGMG---MPPMPPFGMP 1701
PP PG GG G GGG PPPP GGG G PP P G P
Sbjct: 370 PPPPGGGGNPIGGGGGGGNFFFGGGGGGGVPPPPRGGGGGGDPPPPPPAGGAP 528
Score = 36.2 bits (82), Expect = 0.11
Identities = 24/54 (44%), Positives = 26/54 (47%), Gaps = 18/54 (33%)
Frame = +2
Query: 1667 PPMPGMG---------------GGMGGGYAPPPPMGGGMG---MPPMPPFGMPM 1702
PP PG G GG GGG +PPPP GGG G PP PP G P+
Sbjct: 371 PPPPGGGETPSGGGGGGGIFFLGGGGGGGSPPPPGGGGGGGTPPPP-PPRGGPL 529
Score = 34.7 bits (78), Expect = 0.33
Identities = 16/35 (45%), Positives = 17/35 (47%)
Frame = -1
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
PP P GG GG PPPP PP P +G P
Sbjct: 493 PPPPPPPGGGGGPPPPPPPQKKNSPPPPPPRWGFP 389
Score = 34.3 bits (77), Expect = 0.43
Identities = 17/31 (54%), Positives = 18/31 (57%)
Frame = -2
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
PP G GGG+ PPPP GGG PP PP
Sbjct: 522 PPRGGGGGGVPP--PPPPPGGGGDPPPPPPP 436
Score = 33.5 bits (75), Expect = 0.73
Identities = 17/37 (45%), Positives = 17/37 (45%)
Frame = -3
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
P PP P GGG PPPP PP PP G P
Sbjct: 491 PPPPPPRGGGGTP---PPPPPPKKKFPPPPPPPMGFP 390
Score = 30.4 bits (67), Expect = 6.2
Identities = 14/22 (63%), Positives = 14/22 (63%)
Frame = -1
Query: 1683 PPPMGGGMGMPPMPPFGMPMGG 1704
PPP GGG G PP PP P GG
Sbjct: 523 PPPRGGGGGGPPPPP---PPGG 467
Score = 30.4 bits (67), Expect = 6.2
Identities = 20/60 (33%), Positives = 24/60 (39%), Gaps = 22/60 (36%)
Frame = -3
Query: 1661 PLALPAPPMPGMGGGM---------------GGGY-------APPPPMGGGMGMPPMPPF 1698
P + PP PG GGG+ GGG+ PPPP G P+PPF
Sbjct: 410 PPPMGFPPPPGGGGGVFFFCGGGGGPKKKKRGGGFFFFFFDPPPPPPPKKKRGGGPLPPF 231
>TC78027 similar to PIR|D86327|D86327 protein F18O14.17 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 2226
Score = 37.0 bits (84), Expect(2) = 0.012
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = +2
Query: 942 DDELINVTNKNSLFKLQARYVVE 964
D EL+N TN+NS FKL ARYVVE
Sbjct: 1271 DAELVNATNQNSWFKLLARYVVE 1339
Score = 21.2 bits (43), Expect(2) = 0.012
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = +1
Query: 989 VVSTALPESKSPEQVS 1004
VVSTA PESK Q++
Sbjct: 1393 VVSTAFPESKRMLQLN 1440
>TC78025
Length = 1947
Score = 37.0 bits (84), Expect(2) = 0.013
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = +3
Query: 942 DDELINVTNKNSLFKLQARYVVE 964
D EL+N TN+NS FKL ARYVVE
Sbjct: 1101 DAELVNATNQNSWFKLLARYVVE 1169
Score = 21.2 bits (43), Expect(2) = 0.013
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = +2
Query: 989 VVSTALPESKSPEQVS 1004
VVSTA PESK Q++
Sbjct: 1223 VVSTAFPESKRMLQLN 1270
>AL382998 similar to GP|23491723|db formin homology protein A {Dictyostelium
discoideum}, partial (3%)
Length = 355
Score = 37.7 bits (86), Expect = 0.039
Identities = 22/47 (46%), Positives = 22/47 (46%), Gaps = 12/47 (25%)
Frame = +2
Query: 1667 PPMPGMGGGM----GGGYAPPPPMGGGMGMPP--------MPPFGMP 1701
PP P G G GGG APPPP GGG PP PP G P
Sbjct: 2 PPPPPRGRGKKTKKGGGGAPPPPRGGGGAPPPSQKK*GGGSPPRGSP 142
Score = 35.0 bits (79), Expect = 0.25
Identities = 18/30 (60%), Positives = 20/30 (66%), Gaps = 3/30 (10%)
Frame = -2
Query: 1678 GGYAPPPPMGGGMGMPPMP---PFGMPMGG 1704
GG APPPP GGG G PP P F +P+GG
Sbjct: 96 GGGAPPPPRGGG-GAPPPPFFVFFPLPLGG 10
Score = 35.0 bits (79), Expect = 0.25
Identities = 20/46 (43%), Positives = 23/46 (49%), Gaps = 7/46 (15%)
Frame = +1
Query: 1667 PPMPGMG----GGMGGGYAPPPPMGGGMGMPPMPP---FGMPMGGY 1705
PP P G GG PPPP GGG G PP+P G+P G+
Sbjct: 1 PPPPPKGEREENKERGGGGPPPPPGGGGGPPPLPKKIGGGVPPPGF 138
Score = 35.0 bits (79), Expect = 0.25
Identities = 19/40 (47%), Positives = 21/40 (52%), Gaps = 13/40 (32%)
Frame = -1
Query: 1673 GGG------MGGGYAPPPPMGGGMGMP-------PMPPFG 1699
GGG +GGG PPPP GGG G P P PP+G
Sbjct: 130 GGGPPPLFFLGGGGGPPPPPGGGGGPPPPFLCFLPSPPWG 11
Score = 34.7 bits (78), Expect = 0.33
Identities = 19/35 (54%), Positives = 19/35 (54%), Gaps = 2/35 (5%)
Frame = +3
Query: 1665 PAPPMPGMGGGM--GGGYAPPPPMGGGMGMPPMPP 1697
P PP G G GGG PPPP GG G PP PP
Sbjct: 3 PPPPQGGEGRKQRKGGGGPPPPP--GGGGGPPPPP 101
Score = 34.7 bits (78), Expect = 0.33
Identities = 25/63 (39%), Positives = 27/63 (42%), Gaps = 18/63 (28%)
Frame = -3
Query: 1660 LPLALPAPPM------PGMGGG-------MGGGYAPPPPMGGGMGMPPMPPFGM-----P 1701
LP L PP PG GG G G PPPP GGG G P PP + P
Sbjct: 191 LPFFLKEPPFWGGGKFPGNPGGGTPPPIFFGRGGGPPPPPGGGGG--PPPPLSLFSSLSP 18
Query: 1702 MGG 1704
+GG
Sbjct: 17 LGG 9
>BF636172
Length = 669
Score = 36.6 bits (83), Expect = 0.086
Identities = 22/40 (55%), Positives = 26/40 (65%), Gaps = 8/40 (20%)
Frame = +1
Query: 664 QALVEFFGTLSKEWALECMKDLLLV--------NLRGNLQ 695
Q L EFFGTLSKE A E M+DLLLV N+RG+ +
Sbjct: 427 QVLAEFFGTLSKE*APE*MEDLLLVIRNMQSFYNIRGSFK 546
>TC78926 similar to SP|P93231|VP41_LYCES Vacuolar assembly protein VPS41
homolog. [Tomato] {Lycopersicon esculentum}, partial
(39%)
Length = 1641
Score = 36.2 bits (82), Expect = 0.11
Identities = 37/185 (20%), Positives = 77/185 (41%)
Frame = +1
Query: 944 ELINVTNKNSLFKLQARYVVERMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQV 1003
E+ + +K++L + VV+ M D V P R+LI PE V
Sbjct: 187 EVFDFIDKHNLHDVIQEKVVQLMMLDCKRAV--PLYIQNRELISP-----------PEVV 327
Query: 1004 SASVKAFMTADLPHELIELLEKIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLD 1063
+ A +D H L L + N +F+ ++ + D ++ ++
Sbjct: 328 KQLLNADNKSDSRHFLHLYLHSLFEVNPHAGKDFH--DMQVELYADYDPKMLLPFLRSSQ 501
Query: 1064 NFDGPQVGEVAVEAELYEEAFAIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVW 1123
++ + E+ ++ +L +E I + +A+ V+++ + I+ AVEF +D +W
Sbjct: 502 HYTLEKAYEICIKRDLMKEQVFILGRMGNAKKALAVIINKLGDIEEAVEFVTMQHDDELW 681
Query: 1124 SQVAK 1128
++ K
Sbjct: 682 EELIK 696
>TC85803 similar to GP|13785471|dbj|BAB43909. phosphoenolpyruvate
carboxykinase {Flaveria pringlei}, partial (95%)
Length = 2351
Score = 35.4 bits (80), Expect = 0.19
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 16/105 (15%)
Frame = +3
Query: 734 LSSSEDPDIHFKYIEAAAKTGQIKE---VERVTRESSFYDPEKTKN--------FLMEAK 782
LS ++PDI A + G + E + TRE + D T+N ++ AK
Sbjct: 1332 LSREKEPDIW-----NAIRFGTVLENVVFDEHTREVDYSDKSVTENTRAAYPIEYIPNAK 1496
Query: 783 LPDARP-----LINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKV 822
LP P ++ CD FG +P ++ M +I GY V
Sbjct: 1497 LPCVGPHPKNVILLACDAFGVLPPVSKLTLAQTMYHFISGYTALV 1631
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 35.0 bits (79), Expect = 0.25
Identities = 14/21 (66%), Positives = 14/21 (66%)
Frame = +2
Query: 1665 PAPPMPGMGGGMGGGYAPPPP 1685
P PP PG GGG GG PPPP
Sbjct: 602 PPPPHPGRGGGGPGGPPPPPP 664
Score = 33.5 bits (75), Expect = 0.73
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = +3
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFG 1699
P P P GGG GG PPPP G PP G
Sbjct: 603 PPPHTPAGGGGGPGGPRPPPPKKTARGGEERPPAG 707
Score = 31.2 bits (69), Expect = 3.6
Identities = 14/31 (45%), Positives = 17/31 (54%)
Frame = -3
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
PP+P +GGG GGG PP +PP P
Sbjct: 451 PPLPCLGGGRGGGGGAPP-------LPPPSP 380
Score = 30.8 bits (68), Expect = 4.7
Identities = 17/33 (51%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Frame = -1
Query: 1663 ALPAPPMPGMGGGMGGGYAPPPPMGG-GMGMPP 1694
ALP + +GGG GG PPPP G G G PP
Sbjct: 693 ALPPLSLFFLGGGGGGPPGPPPPRPGCGGGGPP 595
>TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 protein
{Arabidopsis thaliana}, partial (87%)
Length = 814
Score = 35.0 bits (79), Expect = 0.25
Identities = 18/43 (41%), Positives = 22/43 (50%)
Frame = -3
Query: 1660 LPLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPM 1702
LPL L PP P +GG P PP+GG + P PP P+
Sbjct: 653 LPLPLKPPPRPPLGG-------PLPPLGGPLPRKPPPPRPPPL 546
>TC93037 weakly similar to GP|3004934|gb|AAC09237.1| annexin XIV {Neurospora
crassa}, partial (33%)
Length = 778
Score = 35.0 bits (79), Expect = 0.25
Identities = 22/54 (40%), Positives = 22/54 (40%), Gaps = 4/54 (7%)
Frame = +3
Query: 1656 YAQLLPLALPAPPMPGMGG----GMGGGYAPPPPMGGGMGMPPMPPFGMPMGGY 1705
Y L LA PG G G GY PPP GG G PP P GGY
Sbjct: 21 Y*PALQLAFQVMSYPGQHGYGAQPHGQGYGQPPPQGGYGGYPPPGGQYPPQGGY 182
>TC82284 weakly similar to GP|11119510|gb|AAF37724.2 diaphanous protein
{Entamoeba histolytica}, partial (4%)
Length = 671
Score = 35.0 bits (79), Expect = 0.25
Identities = 20/40 (50%), Positives = 21/40 (52%), Gaps = 3/40 (7%)
Frame = +1
Query: 1661 PLALPAPPMPGMGGGMGGGY---APPPPMGGGMGMPPMPP 1697
PL P PP G+ G G APPPPM G MPP PP
Sbjct: 85 PLPPPPPPPQGLPPGAPMGNPPRAPPPPMSG--SMPPPPP 198
Score = 31.2 bits (69), Expect = 3.6
Identities = 16/42 (38%), Positives = 18/42 (42%), Gaps = 3/42 (7%)
Frame = +1
Query: 1665 PAPPMPGMGGGMGGGYAP---PPPMGGGMGMPPMPPFGMPMG 1703
P PP G G P PPP + PP PP G+P G
Sbjct: 4 PQPPASAPNGTTAPGVPPRPMPPPPQAPLPPPPPPPQGLPPG 129
>TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box protein {Malus x
domestica}, partial (85%)
Length = 2050
Score = 34.7 bits (78), Expect = 0.33
Identities = 18/38 (47%), Positives = 18/38 (47%), Gaps = 6/38 (15%)
Frame = +1
Query: 1665 PAPPMPGMGGGMGGGYAPPPP------MGGGMGMPPMP 1696
P PP GGG GG Y PPP GG G PP P
Sbjct: 1507 PPPPASTGGGGGGGLYYPPPTGGSGSGSGGNYGTPPPP 1620
Score = 32.0 bits (71), Expect = 2.1
Identities = 16/40 (40%), Positives = 16/40 (40%)
Frame = +1
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P P P GGG G Y PPP P PP GG
Sbjct: 1420 PPPSPPSSGGGSGSTYYSPPPPSVYYYSSPPPPASTGGGG 1539
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,450,324
Number of Sequences: 36976
Number of extensions: 645500
Number of successful extensions: 4696
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 3530
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4443
length of query: 1705
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1596
effective length of database: 4,984,343
effective search space: 7955011428
effective search space used: 7955011428
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC142498.7


