

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.6 - phase: 0
(513 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
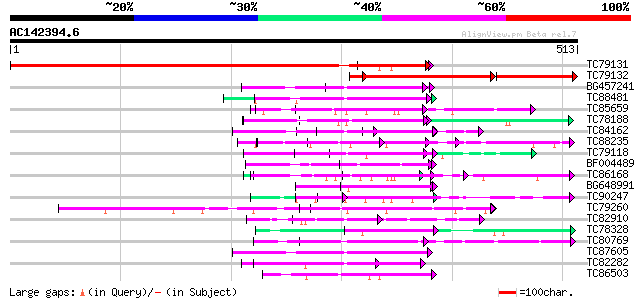
Score E
Sequences producing significant alignments: (bits) Value
TC79131 similar to GP|14031063|gb|AAK52092.1 WD-40 repeat protei... 630 0.0
TC79132 similar to PIR|D96764|D96764 unknown protein F25P22.14 [... 241 e-108
BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [impo... 83 2e-16
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 81 8e-16
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 81 1e-15
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 76 4e-14
TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repea... 73 2e-13
TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Or... 69 3e-12
TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {... 68 7e-12
BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642) ... 68 1e-11
TC86168 homologue to GP|13936812|gb|AAK49947.1 TGF-beta receptor... 65 8e-11
BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] ... 62 4e-10
TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Ara... 62 4e-10
TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat pr... 62 7e-10
TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Ar... 60 3e-09
TC78328 similar to GP|11994333|dbj|BAB02292. WD-40 repeat protei... 59 4e-09
TC80769 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 58 1e-08
TC87605 homologue to PIR|G96563|G96563 probable coatomer complex... 57 2e-08
TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13... 57 2e-08
TC86503 similar to GP|15028127|gb|AAK76687.1 unknown protein {Ar... 56 3e-08
>TC79131 similar to GP|14031063|gb|AAK52092.1 WD-40 repeat protein
{Lycopersicon esculentum}, partial (71%)
Length = 1224
Score = 630 bits (1626), Expect = 0.0
Identities = 332/392 (84%), Positives = 338/392 (85%), Gaps = 12/392 (3%)
Frame = +2
Query: 1 MSTLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAI 60
MSTLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAI
Sbjct: 71 MSTLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAI 250
Query: 61 LPQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEH 120
LPQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEH
Sbjct: 251 LPQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEH 430
Query: 121 LLVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPP 180
LLVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPP
Sbjct: 431 LLVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPP 610
Query: 181 GTQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEV 240
GTQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEV
Sbjct: 611 GTQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEV 790
Query: 241 WDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLR 300
WDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQC
Sbjct: 791 WDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQC-- 964
Query: 301 RLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARI-----------HGLKSGKMLKEFRGHT 349
+ +F R L + + + K +
Sbjct: 965 ------XAAS*ACTFPRCHKCFLLS*WKPIIKYFI*QHCKNPWPQIWKDX*KKYSWAHXX 1126
Query: 350 SYV-NDATFTNDGTRVITASSDCTVKVWDLKT 380
SY NDATFTNDGTRVITASSDCTVKVWDLKT
Sbjct: 1127SYAXNDATFTNDGTRVITASSDCTVKVWDLKT 1222
Score = 41.6 bits (96), Expect = 7e-04
Identities = 24/77 (31%), Positives = 35/77 (45%), Gaps = 8/77 (10%)
Frame = +2
Query: 315 FSRDGSQLLSTSFDSTARIHGLKSGKMLKE--------FRGHTSYVNDATFTNDGTRVIT 366
FS DG L+S S D + SGK+ K+ F H V F+ D + +
Sbjct: 734 FSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIAS 913
Query: 367 ASSDCTVKVWDLKTTDC 383
S+D +KVW ++T C
Sbjct: 914 GSTDGKIKVWRIRTAQC 964
Score = 32.0 bits (71), Expect = 0.59
Identities = 19/73 (26%), Positives = 34/73 (46%), Gaps = 8/73 (10%)
Frame = +2
Query: 448 ACISPKGEWIYCVGEDRNMYCFSYQSGKL--------EHLMTVHDKEIIGVTHHPHRNLV 499
A SP G+++ D + + Y SGKL E +HD+ ++ V ++
Sbjct: 728 ARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMI 907
Query: 500 ATFSDDHTMKLWK 512
A+ S D +K+W+
Sbjct: 908 ASGSTDGKIKVWR 946
>TC79132 similar to PIR|D96764|D96764 unknown protein F25P22.14 [imported] -
Arabidopsis thaliana, partial (36%)
Length = 889
Score = 241 bits (616), Expect(3) = e-108
Identities = 118/120 (98%), Positives = 119/120 (98%)
Frame = +3
Query: 320 SQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK 379
+QLLSTSFDSTARIHGLKSGKM KEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK
Sbjct: 39 AQLLSTSFDSTARIHGLKSGKMWKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK 218
Query: 380 TTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGK 439
TTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGK
Sbjct: 219 TTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGK 398
Score = 164 bits (415), Expect(3) = e-108
Identities = 73/73 (100%), Positives = 73/73 (100%)
Frame = +2
Query: 441 EGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVA 500
EGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVA
Sbjct: 404 EGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVA 583
Query: 501 TFSDDHTMKLWKP 513
TFSDDHTMKLWKP
Sbjct: 584 TFSDDHTMKLWKP 622
Score = 37.7 bits (86), Expect = 0.011
Identities = 39/162 (24%), Positives = 66/162 (40%), Gaps = 12/162 (7%)
Frame = +3
Query: 229 LVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKI 288
L+S S D + SGK+ K+ F H V F+ D + + S+D +
Sbjct: 45 LLSTSFDSTARIHGLKSGKMWKE--------FRGHTSYVNDATFTNDGTRVITASSDCTV 200
Query: 289 KVWRIRTAQCLRRLE-----HAHSQGVTSV-SFSRDGSQLLSTSFDSTARIHGLKSGKML 342
KVW ++T CL+ + V SV F ++ ++ + S+ I L+ G+++
Sbjct: 201 KVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQ-GQVV 377
Query: 343 KEF------RGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
K F R TS +G V+ + CT +L
Sbjct: 378 KSFSSGKTKRAETSLQPAFHLKGNGFTVLVKTGICTASAINL 503
Score = 32.0 bits (71), Expect = 0.59
Identities = 19/73 (26%), Positives = 34/73 (46%)
Frame = +2
Query: 220 ARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMI 279
A SP G+++ D + + Y SGKL E +HD+ ++ V ++
Sbjct: 425 ACISPKGEWIYCVGEDRNMYCFSYQSGKL--------EHLMTVHDKEIIGVTHHPHRNLV 580
Query: 280 ASGSTDGKIKVWR 292
A+ S D +K+W+
Sbjct: 581 ATFSDDHTMKLWK 619
Score = 25.8 bits (55), Expect(3) = e-108
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = +2
Query: 308 QGVTSVSFSRDGSQLL 323
QGVT SFSRDGS ++
Sbjct: 2 QGVTVFSFSRDGSPII 49
>BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [imported] -
Arabidopsis thaliana, partial (15%)
Length = 654
Score = 83.2 bits (204), Expect = 2e-16
Identities = 56/170 (32%), Positives = 79/170 (45%), Gaps = 1/170 (0%)
Frame = +1
Query: 210 KFGAKSH-AECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVL 268
KF KS+ + F P ++++ G I++WDY G L + F HD PV
Sbjct: 82 KFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLI--------DKFDEHDGPVR 237
Query: 269 CVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFD 328
V F + SG D KIKVW + +CL L H + +V F + ++S S D
Sbjct: 238 GVHFHNSQPLFVSGGDDYKIKVWNYKLHRCLFTLL-GHLDYIRTVQFHHESPWIVSASDD 414
Query: 329 STARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
T RI +S + GH YV A F V++AS D TV+VWD+
Sbjct: 415 QTIRIWNWQSRTCISVLTGHNHYVMCALFHPKDDLVVSASLDQTVRVWDI 564
Score = 43.9 bits (102), Expect = 1e-04
Identities = 22/99 (22%), Positives = 44/99 (44%)
Frame = +1
Query: 286 GKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEF 345
G I++W R + + + H V V F +S D ++ K + L
Sbjct: 163 GVIQLWDYRMGTLIDKFDE-HDGPVRGVHFHNSQPLFVSGGDDYKIKVWNYKLHRCLFTL 339
Query: 346 RGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCL 384
GH Y+ F ++ +++AS D T+++W+ ++ C+
Sbjct: 340 LGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCI 456
Score = 41.2 bits (95), Expect = 0.001
Identities = 39/205 (19%), Positives = 72/205 (35%)
Frame = +1
Query: 307 SQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVIT 366
S V +SF +L++ ++ + G ++ +F H V F N ++
Sbjct: 97 SNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFHNSQPLFVS 276
Query: 367 ASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMT 426
D +KVW+ K CL T H+ YI T
Sbjct: 277 GGDDYKIKVWNYKLHRCLFTL----------------------LGHLD--------YIRT 366
Query: 427 LQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKE 486
+Q + WI +D+ + +++QS ++T H+
Sbjct: 367 VQ---------------------FHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHY 483
Query: 487 IIGVTHHPHRNLVATFSDDHTMKLW 511
++ HP +LV + S D T+++W
Sbjct: 484 VMCALFHPKDDLVVSASLDQTVRVW 558
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 81.3 bits (199), Expect = 8e-16
Identities = 47/161 (29%), Positives = 78/161 (48%), Gaps = 1/161 (0%)
Frame = +1
Query: 222 FSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIAS 281
F DG SC +D VWD +G+ L+ H + +L + FS + +A+
Sbjct: 1351 FHHDGSLAASCGLDALARVWDLRTGRSVLALEG--------HVKSILGISFSPNGYHLAT 1506
Query: 282 GSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFS-RDGSQLLSTSFDSTARIHGLKSGK 340
G D ++W +R + L + AHS ++ V F ++G L++ S+D TA++ + K
Sbjct: 1507 GGEDNTCRIWDLRKKKSLYTIP-AHSNLISQVKFEPQEGYFLVTASYDMTAKVWSSRDFK 1683
Query: 341 MLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTT 381
+K GH + V DG ++T S D T+K+W TT
Sbjct: 1684 PVKTLSGHEAKVTSLDVLGDGGYIVTVSHDRTIKLWSXXTT 1806
Score = 69.7 bits (169), Expect = 3e-12
Identities = 53/238 (22%), Positives = 93/238 (38%), Gaps = 45/238 (18%)
Frame = +1
Query: 194 KQDVDDMYPTNLSHTIKFGAKSHAECAR-----------FSPDGQYLVSCSIDGFIEVWD 242
++D D+ + T+K A + E + FS DG+ L +CS G ++W
Sbjct: 856 REDPDEDVDAEIDWTLKQAANLNLEFSEIGDDRPLTGCSFSRDGKGLATCSFTGATKLWS 1035
Query: 243 YISGKLKKDLQYQAEE----------------------------------TFMMHDEPVL 268
+ K L+ + TF H E +
Sbjct: 1036 MPNVKKVSTLKGHTQRATDVAYSPVHKNHLATASADRTAKYWNDQGALLGTFKGHLERLA 1215
Query: 269 CVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFD 328
+ F + + + S D ++W + T + L L+ HS+ V + F DGS S D
Sbjct: 1216 RIAFHPSGKYLGTASYDKTWRLWDVETEEEL-LLQEGHSRSVYGLDFHHDGSLAASCGLD 1392
Query: 329 STARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQT 386
+ AR+ L++G+ + GH + +F+ +G + T D T ++WDL+ L T
Sbjct: 1393 ALARVWDLRTGRSVLALEGHVKSILGISFSPNGYHLATGGEDNTCRIWDLRKKKSLYT 1566
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 80.9 bits (198), Expect = 1e-15
Identities = 52/160 (32%), Positives = 79/160 (48%), Gaps = 4/160 (2%)
Frame = +2
Query: 223 SPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASG 282
S DGQ+ +S S DG + +WD +G + F+ H + VL V FS D+ I S
Sbjct: 275 SSDGQFALSGSWDGELRLWDLNAG--------TSARRFVGHTKDVLSVAFSIDNRQIVSA 430
Query: 283 STDGKIKVWRIRTAQCLRRLE--HAHSQGVTSVSFSRDGSQ--LLSTSFDSTARIHGLKS 338
S D IK+W +C ++ AHS V+ V FS Q ++S S+D T ++ L +
Sbjct: 431 SRDRTIKLWN-TLGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTN 607
Query: 339 GKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
K+ GH+ YVN + DG+ + D + +WDL
Sbjct: 608 CKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDL 727
Score = 65.5 bits (158), Expect = 5e-11
Identities = 61/226 (26%), Positives = 97/226 (41%), Gaps = 9/226 (3%)
Frame = +2
Query: 259 TFMMHDEPVLCVDFSRD-SEMIASGSTDGKIKVWRI----RTAQCLRRLEHAHSQGVTSV 313
T H + V + D S+MI + S D I +W + +T RR HS V V
Sbjct: 89 TMRAHTDVVTAIATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDV 268
Query: 314 SFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTV 373
S DG LS S+D R+ L +G + F GHT V F+ D ++++AS D T+
Sbjct: 269 VLSSDGQFALSGSWDGELRLWDLNAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTI 448
Query: 374 KVWDLKTTDCLQTFKPPPPLRGGDAS---VNSVFIFPKNTE-HIVVCNKTSSIYIMTLQG 429
K+W+ +C T ++ GDA V+ V P + IV + ++ + L
Sbjct: 449 KLWN-TLGECKYT------IQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTN 607
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGK 475
+++ +G G +SP G G+D + + GK
Sbjct: 608 CKLRNTLAG--HSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGK 739
Score = 45.1 bits (105), Expect = 7e-05
Identities = 44/180 (24%), Positives = 79/180 (43%), Gaps = 20/180 (11%)
Frame = +2
Query: 219 CARFSPDGQY--LVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDS 276
C RFSP +VS S D ++VW+ + KL+ T H V V S D
Sbjct: 521 CVRFSPSTLQPTIVSASWDRTVKVWNLTNCKLRN--------TLAGHSGYVNTVAVSPDG 676
Query: 277 EMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGL 336
+ ASG DG I +W + + L L+ + ++ FS + L + + +S+ +I L
Sbjct: 677 SLCASGGKDGVILLWDLAEGKRLYSLDAGSI--IHALCFSPNRYWLCAAT-ESSIKIWDL 847
Query: 337 KSGKMLKEFR------------GHTS------YVNDATFTNDGTRVITASSDCTVKVWDL 378
+S ++++ + G T+ Y ++ DG+ + + +D V+VW +
Sbjct: 848 ESKSIVEDLKVDLKTEADAAIGGDTTTKKKVIYCTSLNWSADGSTLFSGYTDGVVRVWGI 1027
Score = 37.7 bits (86), Expect = 0.011
Identities = 37/142 (26%), Positives = 57/142 (40%), Gaps = 17/142 (11%)
Frame = +2
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G + SPDG S DG I +WD GK Y + ++H LC
Sbjct: 632 GHSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGK----RLYSLDAGSIIH---ALC-- 784
Query: 272 FSRDSEMIASGSTDGKIKVWRIRTAQCLRRLE-------HAHSQG----------VTSVS 314
FS + + + +T+ IK+W + + + L+ A G TS++
Sbjct: 785 FSPNRYWLCA-ATESSIKIWDLESKSIVEDLKVDLKTEADAAIGGDTTTKKKVIYCTSLN 961
Query: 315 FSRDGSQLLSTSFDSTARIHGL 336
+S DGS L S D R+ G+
Sbjct: 962 WSADGSTLFSGYTDGVVRVWGI 1027
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 75.9 bits (185), Expect = 4e-14
Identities = 66/289 (22%), Positives = 113/289 (38%), Gaps = 41/289 (14%)
Frame = +1
Query: 263 HDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQL 322
H V + F+ ++ASGS D +I +W + + H V + ++ DG+Q+
Sbjct: 310 HQSAVYTMKFNPTGSVVASGSHDKEIFLWNVHGDCKNFMVLKGHKNAVLDLHWTSDGTQI 489
Query: 323 LSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG-TRVITASSDCTVKVWDLKTT 381
+S S D T R+ ++GK +K+ H SYVN T G V++ S D T K+WD++
Sbjct: 490 ISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPTRRGPPLVVSGSDDGTAKLWDMRQR 669
Query: 382 DCLQTFKPPPPLRGGDASVNSVFIFPKNTEH-IVVCNKTSSIYIMTLQGQVVKSFSSGKR 440
+QTF + S S I+ ++ + + + MTLQG S
Sbjct: 670 GSIQTFPDKYQITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQGHQDMITSMQLS 849
Query: 441 EGGDFVAA-------CI--------------------------------SPKGEWIYCVG 461
G ++ CI SP G +
Sbjct: 850 PDGSYLLTNGMDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCGWSPDGSKVTAGS 1029
Query: 462 EDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKL 510
DR +Y + S ++ + + H+ + HP+ +V + S D + L
Sbjct: 1030SDRMVYIWDTTSRRILYKLPGHNGSVNECVFHPNEPIVGSCSSDKQIYL 1176
Score = 71.6 bits (174), Expect = 7e-13
Identities = 41/169 (24%), Positives = 81/169 (47%), Gaps = 1/169 (0%)
Frame = +1
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGK-LKKDLQYQAEETFMMHDEPVLCV 270
G K+ ++ DG ++S S D + +WD +GK +KK +++ + C
Sbjct: 436 GHKNAVLDLHWTSDGTQIISASPDKTLRLWDTETGKQIKKMVEHLSYVNS--------CC 591
Query: 271 DFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDST 330
R ++ SGS DG K+W +R ++ + +T+VSFS ++ + D+
Sbjct: 592 PTRRGPPLVVSGSDDGTAKLWDMRQRGSIQTFPDKYQ--ITAVSFSDASDKIYTGGIDND 765
Query: 331 ARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK 379
+I L+ G++ +GH + + DG+ ++T DC + +WD++
Sbjct: 766 VKIWDLRKGEVTMTLQGHQDMITSMQLSPDGSYLLTNGMDCKLCIWDMR 912
Score = 53.1 bits (126), Expect = 2e-07
Identities = 41/178 (23%), Positives = 76/178 (42%), Gaps = 7/178 (3%)
Frame = +1
Query: 211 FGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCV 270
F K FS + + ID +++WD G++ LQ H + + +
Sbjct: 685 FPDKYQITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQG--------HQDMITSM 840
Query: 271 DFSRDSEMIASGSTDGKIKVWRIRTA----QCLRRLE---HAHSQGVTSVSFSRDGSQLL 323
S D + + D K+ +W +R +C++ LE H + + +S DGS++
Sbjct: 841 QLSPDGSYLLTNGMDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCGWSPDGSKVT 1020
Query: 324 STSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTT 381
+ S D I S ++L + GH VN+ F + V + SSD + + ++ +T
Sbjct: 1021AGSSDRMVYIWDTTSRRILYKLPGHNGSVNECVFHPNEPIVGSCSSDKQIYLGEI*ST 1194
>TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repeat protein
{Arabidopsis thaliana}, partial (29%)
Length = 628
Score = 73.2 bits (178), Expect = 2e-13
Identities = 38/110 (34%), Positives = 60/110 (54%)
Frame = +1
Query: 278 MIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLK 337
++ SGS D VWR+ + + H +G+ SV FS +++ S D T RI +
Sbjct: 28 LVCSGSQDRTACVWRLPDLVSVVVFK-GHKRGIWSVEFSPVDQCVVTASGDKTIRIWAIS 204
Query: 338 SGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTF 387
G LK F GHTS V A F GT++++ +D VK+W +K+ +C+ T+
Sbjct: 205 DGSCLKTFEGHTSSVLRALFVTRGTQIVSCGADGLVKLWTVKSNECVATY 354
Score = 52.0 bits (123), Expect = 5e-07
Identities = 32/130 (24%), Positives = 54/130 (40%), Gaps = 1/130 (0%)
Frame = +1
Query: 260 FMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDG 319
F H + V+FS + + + S D I++W I CL+ E H+ V F G
Sbjct: 100 FKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAISDGSCLKTFE-GHTSSVLRALFVTRG 276
Query: 320 SQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVW-DL 378
+Q++S D ++ +KS + + + H V + T SD V +W D
Sbjct: 277 TQIVSCGADGLVKLWTVKSNECVATYDNHEDKVWALAVGRKTETLATGGSDAVVNLWLDS 456
Query: 379 KTTDCLQTFK 388
D + F+
Sbjct: 457 TAADKEEAFR 486
Score = 43.1 bits (100), Expect = 3e-04
Identities = 33/132 (25%), Positives = 53/132 (40%)
Frame = +1
Query: 202 PTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFM 261
P +S + G K FSP Q +V+ S D I +W G K TF
Sbjct: 76 PDLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAISDGSCLK--------TFE 231
Query: 262 MHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQ 321
H VL F I S DG +K+W +++ +C+ ++ H V +++ R
Sbjct: 232 GHTSSVLRALFVTRGTQIVSCGADGLVKLWTVKSNECVATYDN-HEDKVWALAVGRKTET 408
Query: 322 LLSTSFDSTARI 333
L + D+ +
Sbjct: 409 LATGGSDAVVNL 444
Score = 41.6 bits (96), Expect = 7e-04
Identities = 30/109 (27%), Positives = 50/109 (45%)
Frame = +1
Query: 320 SQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK 379
S + S S D TA + L + F+GH + F+ V+TAS D T+++W +
Sbjct: 25 SLVCSGSQDRTACVWRLPDLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAIS 204
Query: 380 TTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQ 428
CL+TF+ G +SV + T+ IV C + + T++
Sbjct: 205 DGSCLKTFE------GHTSSVLRALFVTRGTQ-IVSCGADGLVKLWTVK 330
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/91 (25%), Positives = 39/91 (42%)
Frame = +1
Query: 220 ARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMI 279
A F G +VSC DG +++W S + T+ H++ V + R +E +
Sbjct: 256 ALFVTRGTQIVSCGADGLVKLWTVKSN--------ECVATYDNHEDKVWALAVGRKTETL 411
Query: 280 ASGSTDGKIKVWRIRTAQCLRRLEHAHSQGV 310
A+G +D + +W TA +GV
Sbjct: 412 ATGGSDAVVNLWLDSTAADKEEAFRKEEEGV 504
>TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Oryza
sativa}, partial (91%)
Length = 1080
Score = 69.3 bits (168), Expect = 3e-12
Identities = 63/258 (24%), Positives = 116/258 (44%), Gaps = 16/258 (6%)
Frame = +2
Query: 270 VDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDS 329
+ S+ S ++A+ S D I+ W ++ +C R +++ SQ V + + D + L+ + +
Sbjct: 206 IRMSQPSVLLATASYDHTIRFWEAKSGRCYRTIQYPDSQ-VNRLEITPD-KRYLAAAGNP 379
Query: 330 TARIHGLKSG--KMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTF 387
R+ + S + + + GHTS V F DG + + S D TVK+WDL+ C + +
Sbjct: 380 HIRLFDVNSNSPQPVMSYDGHTSNVMAVGFQCDGNWMYSGSEDGTVKIWDLRAPGCQREY 559
Query: 388 KPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVA 447
+ A+VN+V + P TE ++ ++ +I + L S S D
Sbjct: 560 E-------SRAAVNTVVLHPNQTE-LISGDQNGNIRVWDL---TANSCSCELVPEVDTAV 706
Query: 448 ACISPKGEWIYCVGEDRNMYCFSYQ---------SGKLEHLMTVHDKEIIGVT-----HH 493
++ + V + N C+ ++ + + H + H+ I+
Sbjct: 707 RSLTVMWDGSLVVAANNNGTCYVWRLLRGTQTMTNFEPLHKLQAHNGYILKCVLSPEFCD 886
Query: 494 PHRNLVATFSDDHTMKLW 511
PHR L AT S DHT+K+W
Sbjct: 887 PHRYL-ATASSDHTVKIW 937
Score = 58.9 bits (141), Expect = 4e-09
Identities = 52/217 (23%), Positives = 89/217 (40%), Gaps = 15/217 (6%)
Frame = +2
Query: 207 HTIKFGAKSHAECAR-------------FSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQ 253
HTI+F C R +PD +YL + + I ++D S + +
Sbjct: 254 HTIRFWEAKSGRCYRTIQYPDSQVNRLEITPDKRYLAAAG-NPHIRLFDVNSNSPQPVMS 430
Query: 254 YQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSV 313
Y H V+ V F D + SGS DG +K+W +R C R E+ V +V
Sbjct: 431 YDG------HTSNVMAVGFQCDGNWMYSGSEDGTVKIWDLRAPGCQR--EYESRAAVNTV 586
Query: 314 SFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHT-SYVNDATFTNDGTRVITASSDCT 372
+ ++L+S + R+ L + E + V T DG+ V+ A+++ T
Sbjct: 587 VLHPNQTELISGDQNGNIRVWDLTANSCSCELVPEVDTAVRSLTVMWDGSLVVAANNNGT 766
Query: 373 VKVWD-LKTTDCLQTFKPPPPLRGGDASVNSVFIFPK 408
VW L+ T + F+P L+ + + + P+
Sbjct: 767 CYVWRLLRGTQTMTNFEPLHKLQAHNGYILKCVLSPE 877
Score = 56.2 bits (134), Expect = 3e-08
Identities = 44/167 (26%), Positives = 68/167 (40%), Gaps = 10/167 (5%)
Frame = +2
Query: 224 PDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGS 283
P+ L+S +G I VWD + +L + D V + D ++ + +
Sbjct: 596 PNQTELISGDQNGNIRVWDLTANSCSCELVPEV-------DTAVRSLTVMWDGSLVVAAN 754
Query: 284 TDGKIKVWRI-RTAQCLRRLEHAHS---------QGVTSVSFSRDGSQLLSTSFDSTARI 333
+G VWR+ R Q + E H + V S F L + S D T +I
Sbjct: 755 NNGTCYVWRLLRGTQTMTNFEPLHKLQAHNGYILKCVLSPEFCDPHRYLATASSDHTVKI 934
Query: 334 HGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKT 380
+ + K GH + D F+ DG +ITASSD T ++W T
Sbjct: 935 WNVDDFTLEKTLIGHQRRLWDCVFSVDGAYLITASSDSTARLWSTTT 1075
Score = 42.7 bits (99), Expect = 3e-04
Identities = 31/119 (26%), Positives = 56/119 (47%), Gaps = 4/119 (3%)
Frame = +2
Query: 225 DGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVL-CV---DFSRDSEMIA 280
DG +V+ + +G VW + G + ++ H+ +L CV +F +A
Sbjct: 728 DGSLVVAANNNGTCYVWRLLRGT-QTMTNFEPLHKLQAHNGYILKCVLSPEFCDPHRYLA 904
Query: 281 SGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSG 339
+ S+D +K+W + L + H + + FS DG+ L++ S DSTAR+ +G
Sbjct: 905 TASSDHTVKIWNVDDFT-LEKTLIGHQRRLWDCVFSVDGAYLITASSDSTARLWSTTTG 1078
>TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {Arabidopsis
thaliana}, partial (44%)
Length = 1098
Score = 68.2 bits (165), Expect = 7e-12
Identities = 42/169 (24%), Positives = 78/169 (45%), Gaps = 1/169 (0%)
Frame = +2
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPV-LCV 270
G + C ++ P G L SCS D ++W K D + ++E + + P
Sbjct: 311 GHQGEVNCVKWDPTGTLLASCSDDITAKIWSVKQDKYLHDFREHSKEIYTIRWSPTGPGT 490
Query: 271 DFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDST 330
+ ++AS S D +K+W I + + L + H V SV+FS +G + S S D +
Sbjct: 491 NNPNKKLLLASASFDSTVKLWDIELGKLIHSL-NGHRHPVYSVAFSPNGEYIASGSLDKS 667
Query: 331 ARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK 379
I LK GK+++ + G + + + + +G ++ ++ V V D +
Sbjct: 668 LHIWSLKEGKIIRTYNG-SGGIFEVCWNKEGDKIAACFANNIVCVLDFR 811
Score = 62.8 bits (151), Expect = 3e-10
Identities = 37/139 (26%), Positives = 66/139 (46%), Gaps = 9/139 (6%)
Frame = +2
Query: 258 ETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSR 317
+TF H V CV + ++AS S D K+W ++ + L HS+ + ++ +S
Sbjct: 299 QTFAGHQGEVNCVKWDPTGTLLASCSDDITAKIWSVKQDKYLHDFRE-HSKEIYTIRWSP 475
Query: 318 DGSQ---------LLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITAS 368
G L S SFDST ++ ++ GK++ GH V F+ +G + + S
Sbjct: 476 TGPGTNNPNKKLLLASASFDSTVKLWDIELGKLIHSLNGHRHPVYSVAFSPNGEYIASGS 655
Query: 369 SDCTVKVWDLKTTDCLQTF 387
D ++ +W LK ++T+
Sbjct: 656 LDKSLHIWSLKEGKIIRTY 712
Score = 45.4 bits (106), Expect = 5e-05
Identities = 41/196 (20%), Positives = 80/196 (39%), Gaps = 9/196 (4%)
Frame = +2
Query: 290 VWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHT 349
VW ++ +C ++ HS V + R + S+S D+ + + + F GH
Sbjct: 146 VWDVKAEECKQQFGF-HSGPTLDVDW-RTNTSFASSSTDNMIYVCKIGENHPTQTFAGHQ 319
Query: 350 SYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPP---------PPLRGGDASV 400
VN + GT + + S D T K+W +K L F+ P G +
Sbjct: 320 GEVNCVKWDPTGTLLASCSDDITAKIWSVKQDKYLHDFREHSKEIYTIRWSPTGPGTNNP 499
Query: 401 NSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCV 460
N + + + T ++ + L G+++ S + + + SP GE+I
Sbjct: 500 NKKLLLASAS-----FDSTVKLWDIEL-GKLIHSLNGHRH---PVYSVAFSPNGEYIASG 652
Query: 461 GEDRNMYCFSYQSGKL 476
D++++ +S + GK+
Sbjct: 653 SLDKSLHIWSLKEGKI 700
>BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642)
D41819(S4642)~similar to Arabidopsis thaliana chromosome
1 At1g49040~, partial (16%)
Length = 605
Score = 67.8 bits (164), Expect = 1e-11
Identities = 46/171 (26%), Positives = 78/171 (44%), Gaps = 1/171 (0%)
Frame = +1
Query: 214 KSHAECAR-FSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDF 272
K H R S D +VS S D + VWD + +L ++L+ HD PV CV
Sbjct: 16 KGHTRTVRAISSDMGKVVSGSDDQSVLVWDKQTTQLLEELKG--------HDGPVSCVR- 168
Query: 273 SRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTAR 332
+ E + + S DG +K+W +RT +C+ + S V + + + L + D A
Sbjct: 169 TLSGERVLTASHDGTVKMWDVRTDRCVATVGRC-SSAVLCMEYDDNVGILAAAGRDVVAN 345
Query: 333 IHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDC 383
+ +++ + + + GHT ++ D VIT S D T ++W + C
Sbjct: 346 MWDIRASRQMHKLSGHTQWIRSLRMVGD--TVITGSDDWTARIWSVSRGTC 492
Score = 50.4 bits (119), Expect = 2e-06
Identities = 30/88 (34%), Positives = 51/88 (57%)
Frame = +1
Query: 299 LRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFT 358
LR H++ V ++S D +++S S D + + ++ ++L+E +GH V+ T
Sbjct: 1 LRATLKGHTRTVRAIS--SDMGKVVSGSDDQSVLVWDKQTTQLLEELKGHDGPVS-CVRT 171
Query: 359 NDGTRVITASSDCTVKVWDLKTTDCLQT 386
G RV+TAS D TVK+WD++T C+ T
Sbjct: 172 LSGERVLTASHDGTVKMWDVRTDRCVAT 255
Score = 34.7 bits (78), Expect = 0.090
Identities = 18/66 (27%), Positives = 29/66 (43%)
Frame = +1
Query: 226 GQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTD 285
G +++ S D +W G + H P+LCV++S I +GSTD
Sbjct: 424 GDTVITGSDDWTARIWSVSRGT--------CDAVLACHAGPILCVEYSSLDRGIITGSTD 579
Query: 286 GKIKVW 291
G ++ W
Sbjct: 580 GLLRFW 597
>TC86168 homologue to GP|13936812|gb|AAK49947.1 TGF-beta
receptor-interacting protein 1 {Phaseolus vulgaris},
complete
Length = 1322
Score = 64.7 bits (156), Expect = 8e-11
Identities = 56/217 (25%), Positives = 90/217 (40%), Gaps = 7/217 (3%)
Frame = +2
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
L H + +T + ++RDG L S + D + +G+ L +RGH V + D
Sbjct: 182 LMKGHERPLTFLKYNRDGDLLFSCAKDHNPTVWFADNGERLGTYRGHNGAVWCCDVSRDS 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
R+IT S+D T K+W+++T L TF P R D SV + + +S+
Sbjct: 362 GRLITGSADQTAKLWNVQTGQQLFTFNFDSPARSVDFSVGDKLAVITTDPFMEL---SSA 532
Query: 422 IYIMTL----QGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKL- 476
I++ + Q +S K G A P + I GED + + ++GKL
Sbjct: 533 IHVKRIAKDPSDQTAESLLVIKGPQGRINRAIWGPLNKTIISAGEDAVIRIWDSETGKLL 712
Query: 477 --EHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
H K I + + T S D + K+W
Sbjct: 713 KESDKELGHKKTITSLAKSADGSHFCTGSLDKSAKIW 823
Score = 54.3 bits (129), Expect = 1e-07
Identities = 45/176 (25%), Positives = 74/176 (41%), Gaps = 11/176 (6%)
Frame = +2
Query: 221 RFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIA 280
+++ DG L SC+ D VW +G + T+ H+ V C D SRDS +
Sbjct: 218 KYNRDGDLLFSCAKDHNPTVWFADNG--------ERLGTYRGHNGAVWCCDVSRDSGRLI 373
Query: 281 SGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTS---FDSTARIHGLK 337
+GS D K+W ++T Q L SV FS + T+ + ++ IH +
Sbjct: 374 TGSADQTAKLWNVQTGQQL--FTFNFDSPARSVDFSVGDKLAVITTDPFMELSSAIHVKR 547
Query: 338 SGK--------MLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQ 385
K L +G +N A + +I+A D +++WD +T L+
Sbjct: 548 IAKDPSDQTAESLLVIKGPQGRINRAIWGPLNKTIISAGEDAVIRIWDSETGKLLK 715
Score = 47.4 bits (111), Expect = 1e-05
Identities = 52/212 (24%), Positives = 90/212 (41%), Gaps = 15/212 (7%)
Frame = +2
Query: 219 CARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEM 278
C S D L++ S D ++W+ +G+ + D P VDFS +
Sbjct: 338 CCDVSRDSGRLITGSADQTAKLWNVQTGQQLFTFNF---------DSPARSVDFSV-GDK 487
Query: 279 IASGSTD------GKIKVWRI------RTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTS 326
+A +TD I V RI +TA+ L ++ + + + ++S
Sbjct: 488 LAVITTDPFMELSSAIHVKRIAKDPSDQTAESLLVIKGPQGR-INRAIWGPLNKTIISAG 664
Query: 327 FDSTARIHGLKSGKMLKEFR---GHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDC 383
D+ RI ++GK+LKE GH + + DG+ T S D + K+WD +T
Sbjct: 665 EDAVIRIWDSETGKLLKESDKELGHKKTITSLAKSADGSHFCTGSLDKSAKIWDSRTLSL 844
Query: 384 LQTFKPPPPLRGGDASVNSVFIFPKNTEHIVV 415
++T+ P VN+V + P +H+V+
Sbjct: 845 VKTYVTERP-------VNAVTMSPL-LDHVVL 916
Score = 42.4 bits (98), Expect = 4e-04
Identities = 39/179 (21%), Positives = 71/179 (38%), Gaps = 15/179 (8%)
Frame = +2
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G + A + P + ++S D I +WD +GKL K+ + + H + + +
Sbjct: 599 GPQGRINRAIWGPLNKTIISAGEDAVIRIWDSETGKLLKE-----SDKELGHKKTITSLA 763
Query: 272 FSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFS-----------RDGS 320
S D +GS D K+W RT ++ + + V +V+ S +D S
Sbjct: 764 KSADGSHFCTGSLDKSAKIWDSRTLSLVK--TYVTERPVNAVTMSPLLDHVVLGGGQDAS 937
Query: 321 QLLSTSFDSTARIHGLKSGKMLKE----FRGHTSYVNDATFTNDGTRVITASSDCTVKV 375
+ +T + K+L+E +GH +N F DG + D V++
Sbjct: 938 AVTTTD-HRAGKFEAKFFDKILQEEIGGVKGHFGPINALAFNPDGKSFSSGGEDGYVRL 1111
>BG648991 similar to PIR|C86239|C86 protein T10O24.21 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 702
Score = 62.4 bits (150), Expect = 4e-10
Identities = 32/93 (34%), Positives = 53/93 (56%), Gaps = 5/93 (5%)
Frame = +3
Query: 300 RRLEHA---HSQGVTSVSF-SRDGSQLLSTSFDSTARIHGL-KSGKMLKEFRGHTSYVND 354
+RL H H++GV+++ F G +LS D+ +I + +GK ++ + GH+ V D
Sbjct: 297 KRLVHTWSGHTKGVSAIRFFPNSGHLILSAGMDTKVKIWDVFNTGKCMRTYMGHSKAVRD 476
Query: 355 ATFTNDGTRVITASSDCTVKVWDLKTTDCLQTF 387
FTNDGT+ ++A D +K WD +T + TF
Sbjct: 477 ICFTNDGTKFLSAGYDKNIKYWDTETGQVISTF 575
Score = 54.7 bits (130), Expect = 8e-08
Identities = 37/131 (28%), Positives = 62/131 (47%), Gaps = 4/131 (3%)
Frame = +3
Query: 259 TFMMHDEPVLCVDFSRDS-EMIASGSTDGKIKVWRI-RTAQCLRRLEHAHSQGVTSVSFS 316
T+ H + V + F +S +I S D K+K+W + T +C+R HS+ V + F+
Sbjct: 312 TWSGHTKGVSAIRFFPNSGHLILSAGMDTKVKIWDVFNTGKCMRTY-MGHSKAVRDICFT 488
Query: 317 RDGSQLLSTSFDSTARIHGLKSGKMLKEF-RGHTSYVNDATFTNDGTRVITAS-SDCTVK 374
DG++ LS +D + ++G+++ F G YV D V+ A SD +
Sbjct: 489 NDGTKFLSAGYDKNIKYWDTETGQVISTFSTGKIPYVVKLNPDEDKQNVLLAGMSDKKIV 668
Query: 375 VWDLKTTDCLQ 385
WD+ T Q
Sbjct: 669 QWDMNTGQITQ 701
>TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Arabidopsis
thaliana}, partial (60%)
Length = 1041
Score = 62.4 bits (150), Expect = 4e-10
Identities = 62/246 (25%), Positives = 102/246 (41%), Gaps = 13/246 (5%)
Frame = +1
Query: 279 IASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGL-- 336
+ G+ G I +W + T + L++ HAH + VT + FS D S L+S S D R+ L
Sbjct: 361 LVGGALSGDIYLWEVETGRLLKKW-HAHLRAVTCLVFSDDDSLLISGSEDGYVRVWSLFS 537
Query: 337 -------KSGKMLKE--FRGHTSYVNDATFTNDGTR--VITASSDCTVKVWDLKTTDCLQ 385
+ K L E F HT V D N G +++AS D T KVW L L+
Sbjct: 538 LFDDLRSREEKNLYEYSFSEHTMRVTDVVIGNGGCNAIIVSASDDRTCKVWSLSRGTLLR 717
Query: 386 TFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDF 445
+ P + +N++ + P EH+ F +G +G F
Sbjct: 718 SIVFP-------SVINAIVLDP--AEHV---------------------FYAGSSDGKIF 807
Query: 446 VAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDD 505
+AA + + I ++ M+ S H K + + + RNL+ + S+D
Sbjct: 808 IAALNTERITTI----DNYGMHIIGSFSN--------HSKAVTCLAYSKGRNLLISGSED 951
Query: 506 HTMKLW 511
+++W
Sbjct: 952 GIVRVW 969
Score = 50.8 bits (120), Expect = 1e-06
Identities = 52/194 (26%), Positives = 79/194 (39%), Gaps = 24/194 (12%)
Frame = +1
Query: 219 CARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEE-----TFMMHDEPVLCVDFS 273
C FS D L+S S DG++ VW S L DL+ + E+ +F H V V
Sbjct: 457 CLVFSDDDSLLISGSEDGYVRVWSLFS--LFDDLRSREEKNLYEYSFSEHTMRVTDVVIG 630
Query: 274 RD--SEMIASGSTDGKIKVWRIRTAQCLRRL--------------EHAHSQGVTSVSFSR 317
+ +I S S D KVW + LR + EH G S
Sbjct: 631 NGGCNAIIVSASDDRTCKVWSLSRGTLLRSIVFPSVINAIVLDPAEHVFYAG------SS 792
Query: 318 DGS---QLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVK 374
DG L+T +T +G+ ++ F H+ V ++ +I+ S D V+
Sbjct: 793 DGKIFIAALNTERITTIDNYGM---HIIGSFSNHSKAVTCLAYSKGRNLLISGSEDGIVR 963
Query: 375 VWDLKTTDCLQTFK 388
VW+ KT + ++ FK
Sbjct: 964 VWNAKTHNIVRMFK 1005
Score = 41.6 bits (96), Expect = 7e-04
Identities = 16/47 (34%), Positives = 28/47 (59%)
Frame = +1
Query: 259 TFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHA 305
+F H + V C+ +S+ ++ SGS DG ++VW +T +R +HA
Sbjct: 871 SFSNHSKAVTCLAYSKGRNLLISGSEDGIVRVWNAKTHNIVRMFKHA 1011
>TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-like
{Arabidopsis thaliana}, partial (50%)
Length = 1227
Score = 61.6 bits (148), Expect = 7e-10
Identities = 46/185 (24%), Positives = 90/185 (47%), Gaps = 8/185 (4%)
Frame = +2
Query: 263 HDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEH---AHSQGVTSVSFSRDG 319
HD+ V V FS D + +A+ S D +W + T L ++H H + V+SVS+S +G
Sbjct: 542 HDDEVWFVQFSHDGKYLATASNDRTAIIWEVDTNDGL-SMKHKLSGHQKSVSSVSWSPNG 718
Query: 320 SQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK 379
+LL+ + R + +GK L+ + + S + + G +++ SD ++ +W+L
Sbjct: 719 QELLTCGVEEAVRRWDVSTGKCLQVYEKNGSGLISCAWFPCGKYILSGLSDKSICMWELD 898
Query: 380 TTDCLQTFKPPPPLRGGDASVNS-----VFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKS 434
+ ++++K L+ D + + I +N I++ NK + + + Q + S
Sbjct: 899 GKE-VESWKGQKTLKISDLEITGDGEHILSICKENA--ILLFNKETKVERFIEEDQTITS 1069
Query: 435 FSSGK 439
FS K
Sbjct: 1070FSLSK 1084
Score = 50.8 bits (120), Expect = 1e-06
Identities = 49/171 (28%), Positives = 79/171 (45%), Gaps = 4/171 (2%)
Frame = +2
Query: 273 SRDSEM-IASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTA 331
S D EM + S GK ++ RT Q L AH V V FS DG L + S D TA
Sbjct: 455 SLDKEMSLYSDHHCGKTQI-PSRTLQIL----DAHDDEVWFVQFSHDGKYLATASNDRTA 619
Query: 332 RIHGLKSG---KMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFK 388
I + + M + GH V+ +++ +G ++T + V+ WD+ T CLQ ++
Sbjct: 620 IIWEVDTNDGLSMKHKLSGHQKSVSSVSWSPNGQELLTCGVEEAVRRWDVSTGKCLQVYE 799
Query: 389 PPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGK 439
+ G ++ + FP ++I+ SI + L G+ V+S+ K
Sbjct: 800 -----KNGSGLISCAW-FPCG-KYILSGLSDKSICMWELDGKEVESWKGQK 931
Score = 47.4 bits (111), Expect = 1e-05
Identities = 76/411 (18%), Positives = 167/411 (40%), Gaps = 15/411 (3%)
Frame = +2
Query: 45 IETFVADINGGRWDAILPQVSQLKLPRNTLEDLYEQIVLEM--IELRELDTARAILRQTQ 102
+ F+ + G W+ + ++++ L + ++LE EL + D L+ +
Sbjct: 32 VNVFMQQVLDGDWEESVSTLNKIGLEDERIVRAASFLILEQKFFELLDGDKVMDALKTLR 211
Query: 103 VMGVMKQEQPERYLRLEHLLVRTYFDPNEAYQESTKEKRRAQIA---QAIAAEVTVVPPS 159
R L L+ + ++ + K R ++ Q + ++P +
Sbjct: 212 TEITELCVDSARIRELSSCLLSPSDQCGSSGRDFIRAKTRLKLLEELQKLIPPTVMIPEN 391
Query: 160 RLMALIGQALKWQQ-----HQGLLPPGTQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAK 214
RL L+ QAL Q+ H L + L+ K + S T++
Sbjct: 392 RLEHLVEQALILQRDACSFHNSL---DKEMSLYSDHHCGKTQIP-------SRTLQILDA 541
Query: 215 SHAEC--ARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDF 272
E +FS DG+YL + S D +W+ + + + H + V V +
Sbjct: 542 HDDEVWFVQFSHDGKYLATASNDRTAIIWE-----VDTNDGLSMKHKLSGHQKSVSSVSW 706
Query: 273 SRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTAR 332
S + + + + + ++ W + T +CL+ E + G+ S ++ G +LS D +
Sbjct: 707 SPNGQELLTCGVEEAVRRWDVSTGKCLQVYE-KNGSGLISCAWFPCGKYILSGLSDKSIC 883
Query: 333 IHGLKSGKMLKEFRGH-TSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPP 391
+ L GK ++ ++G T ++D T DG +++ + + +++ +T ++ F
Sbjct: 884 MWEL-DGKEVESWKGQKTLKISDLEITGDGEHILSICKENAILLFNKETK--VERFIEE- 1051
Query: 392 PLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG--QVVKSFSSGKR 440
D ++ S F K++ ++V I++ ++G ++V + S +R
Sbjct: 1052-----DQTITS-FSLSKDSRFLLVNLLNQEIHLWNIEGDLKLVGKYKSHRR 1186
>TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Arabidopsis
thaliana}, partial (19%)
Length = 821
Score = 59.7 bits (143), Expect = 3e-09
Identities = 35/123 (28%), Positives = 61/123 (49%)
Frame = +1
Query: 215 SHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSR 274
S EC F+P G + +D + +WD +++ + H+ V C +
Sbjct: 172 SSIECVGFAPSGSWAAIGGMDK-MTIWD---------VEHSLARSICEHEYGVTCSTWLG 321
Query: 275 DSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIH 334
S +A+GS DG +++W R+ +C+R HS+ + S+S S + L+S S D TAR+
Sbjct: 322 TS-YVATGSNDGAVRLWDSRSGECVRTFR-GHSESIQSLSLSANQEYLVSASLDHTARVF 495
Query: 335 GLK 337
+K
Sbjct: 496 DVK 504
Score = 47.0 bits (110), Expect = 2e-05
Identities = 43/179 (24%), Positives = 82/179 (45%), Gaps = 6/179 (3%)
Frame = +1
Query: 257 EETFMM----HDEP--VLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGV 310
E+ FM+ H P + C+ + S + +GS DG + + I T + + L +HS +
Sbjct: 4 EKAFMLCEVIHTIPRGITCLAINSTSTIALTGSVDGSVHIVNITTGRVVSTLP-SHSSSI 180
Query: 311 TSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSD 370
V F+ GS D I ++ + + H V +T+ + V T S+D
Sbjct: 181 ECVGFAPSGSWAAIGGMDKMT-IWDVEHS-LARSICEHEYGVTCSTWLGT-SYVATGSND 351
Query: 371 CTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG 429
V++WD ++ +C++TF RG S+ S+ + N E++V + + + ++G
Sbjct: 352 GAVRLWDSRSGECVRTF------RGHSESIQSLSL-SANQEYLVSASLDHTARVFDVKG 507
>TC78328 similar to GP|11994333|dbj|BAB02292. WD-40 repeat protein-like
{Arabidopsis thaliana}, partial (88%)
Length = 1516
Score = 58.9 bits (141), Expect = 4e-09
Identities = 70/300 (23%), Positives = 113/300 (37%), Gaps = 10/300 (3%)
Frame = +1
Query: 223 SPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASG 282
+PDG +L+S S D S L+ TF H V ++ A+
Sbjct: 343 TPDGFFLISASKDA--------SPMLRNGETGDWIGTFEGHKGAVWSCCLDANALRAATA 498
Query: 283 STDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGL-KSGKM 341
S D KVW T L EH H V + +FS D LL+ + RI+ L +
Sbjct: 499 SADFSTKVWDALTGDALHSFEHKHI--VRACAFSEDTHLLLTGGAEKILRIYDLNRPDAP 672
Query: 342 LKEFRGHTSYVNDATFTNDGTRVITASSDC-TVKVWDLKTTDCLQTFKPPPPLRGGDASV 400
KE V + + ++++ +D V++WD++T +QT P+ + S
Sbjct: 673 PKEVDKSPGSVRSVAWLHSDQTILSSCTDMGGVRLWDVRTGKIVQTLDTKSPMTSAEVS- 849
Query: 401 NSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSS---GKREGGDF----VAACISPK 453
YI T G VK + + G + D +A + PK
Sbjct: 850 ------------------QDGRYITTADGSTVKFWDANHYGLVKSYDMPCNVESASLEPK 975
Query: 454 -GEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLWK 512
G GED ++ F + +G H + V P A+ S+D T+++W+
Sbjct: 976 FGNKFVAGGEDLWVHVFDFHTGDEIACNKGHHGPVHCVRFSPGGESYASGSEDGTIRIWQ 1155
Score = 50.8 bits (120), Expect = 1e-06
Identities = 29/88 (32%), Positives = 44/88 (49%), Gaps = 3/88 (3%)
Frame = +1
Query: 304 HAHSQGVTSVSFSR---DGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTND 360
H HS+ V + +S DG L+S S D++ + ++G + F GH V +
Sbjct: 298 HGHSRPVVDLFYSPVTPDGFFLISASKDASPMLRNGETGDWIGTFEGHKGAVWSCCLDAN 477
Query: 361 GTRVITASSDCTVKVWDLKTTDCLQTFK 388
R TAS+D + KVWD T D L +F+
Sbjct: 478 ALRAATASADFSTKVWDALTGDALHSFE 561
Score = 37.4 bits (85), Expect = 0.014
Identities = 20/49 (40%), Positives = 26/49 (52%)
Frame = +1
Query: 263 HDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVT 311
H PV CV FS E ASGS DG I++W+ L H S+G++
Sbjct: 1066 HHGPVHCVRFSPGGESYASGSEDGTIRIWQTGP------LAHDESEGLS 1194
Score = 30.4 bits (67), Expect = 1.7
Identities = 16/40 (40%), Positives = 19/40 (47%)
Frame = +1
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKD 251
G C RFSP G+ S S DG I +W +G L D
Sbjct: 1063 GHHGPVHCVRFSPGGESYASGSEDGTIRIWQ--TGPLAHD 1176
>TC80769 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (40%)
Length = 1362
Score = 57.8 bits (138), Expect = 1e-08
Identities = 59/251 (23%), Positives = 103/251 (40%), Gaps = 1/251 (0%)
Frame = +3
Query: 263 HDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQL 322
H + V FS +A+ S D ++VW + R HS V S+ F + L
Sbjct: 489 HSALITDVRFSASMPRLATSSFDKTVRVWDVDNPGYSLRTFTGHSTSVMSLDFHPNKDDL 668
Query: 323 L-STSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTT 381
+ S D R + +G ++ +G T+ + F R + A+++ V + D++T
Sbjct: 669 ICSCDGDGEIRYWSINNGSCVRVSKGGTTQMR---FQPRLGRYLAAAAENIVSILDVETQ 839
Query: 382 DCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKRE 441
C + L+G +++SV P ++ S+ I TL+G+ V S
Sbjct: 840 ACRYS------LKGHTKTIDSVCWDPSG--ELLASVSEDSVRIWTLEGECVHELSCN--- 986
Query: 442 GGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVAT 501
G F + P + +G +++ + K L + HD I + LVA+
Sbjct: 987 GSKFHSCVFHPTFPSLLVIGCYQSLELCNMAENKTMTL-SAHDGLITALAVSTVNGLVAS 1163
Query: 502 FSDDHTMKLWK 512
S D +KLWK
Sbjct: 1164ASHDKFIKLWK 1196
Score = 53.9 bits (128), Expect = 1e-07
Identities = 39/160 (24%), Positives = 69/160 (42%), Gaps = 1/160 (0%)
Frame = +3
Query: 221 RFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSE-MI 279
RFS L + S D + VWD D + TF H V+ +DF + + +I
Sbjct: 513 RFSASMPRLATSSFDKTVRVWDV-------DNPGYSLRTFTGHSTSVMSLDFHPNKDDLI 671
Query: 280 ASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSG 339
S DG+I+ W I C+R G T + F + L+ + ++ I +++
Sbjct: 672 CSCDGDGEIRYWSINNGSCVR----VSKGGTTQMRFQPRLGRYLAAAAENIVSILDVETQ 839
Query: 340 KMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLK 379
+GHT ++ + G + + S D +V++W L+
Sbjct: 840 ACRYSLKGHTKTIDSVCWDPSGELLASVSED-SVRIWTLE 956
>TC87605 homologue to PIR|G96563|G96563 probable coatomer complex subunit
33791-27676 [imported] - Arabidopsis thaliana, partial
(23%)
Length = 765
Score = 57.0 bits (136), Expect = 2e-08
Identities = 45/184 (24%), Positives = 82/184 (44%), Gaps = 3/184 (1%)
Frame = +3
Query: 202 PTNLSHTIKFGAKSH-AECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETF 260
P L K +S +C P ++++ G + +W+Y S + K +F
Sbjct: 129 PLRLEIKRKLAQRSERVKCVDLHPTEPWILASLYSGTVCIWNYQSQTMAK--------SF 284
Query: 261 MMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGS 320
+ + PV F + + +G+ D I+V+ T ++ E AH+ + V+
Sbjct: 285 EVTELPVRSAKFVARKQWVVAGADDMFIRVYNYNTMDKVKVFE-AHTDYIRCVAVHPTLP 461
Query: 321 QLLSTSFDSTARIHGLKSGKMLKE-FRGHTSYVNDATFTNDGTRVI-TASSDCTVKVWDL 378
+LS+S D ++ + G + + F GH+ YV TF T +AS D T+K+W+L
Sbjct: 462 YVLSSSDDMLIKLWDWEKGWICTQIFEGHSHYVMQVTFNPKDTNTFASASLDRTIKIWNL 641
Query: 379 KTTD 382
+ D
Sbjct: 642 GSPD 653
Score = 29.3 bits (64), Expect = 3.8
Identities = 15/57 (26%), Positives = 25/57 (43%)
Frame = +3
Query: 455 EWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
+W+ +D + ++Y + + H I V HP V + SDD +KLW
Sbjct: 333 QWVVAGADDMFIRVYNYNTMDKVKVFEAHTDYIRCVAVHPTLPYVLSSSDDMLIKLW 503
>TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13.70 -
Arabidopsis thaliana, partial (84%)
Length = 904
Score = 57.0 bits (136), Expect = 2e-08
Identities = 48/163 (29%), Positives = 73/163 (44%), Gaps = 7/163 (4%)
Frame = +2
Query: 221 RFSPDGQYL-VSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEP-----VLCVDFSR 274
RF P G L V+ + +WD + +L L E D+ VL V +S
Sbjct: 401 RFDPKGAILAVAGGGSASVNLWDTSTWELVVTLSIPRVEGPKPSDKSGSKKFVLSVAWSP 580
Query: 275 DSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSR-DGSQLLSTSFDSTARI 333
D + +A GS DG I V+ ++ A+ L LE H V S+ +S D L S S D +
Sbjct: 581 DGKRLACGSMDGTISVFDVQRAKFLHHLE-GHFMPVRSLVYSPYDPRLLFSASDDGNVHM 757
Query: 334 HGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVW 376
+ + + H S+V + +G + T SSD TV++W
Sbjct: 758 YDAEGKALXGTMSXHASWVLCVDVSPNGAAIATGSSDKTVRLW 886
Score = 44.3 bits (103), Expect = 1e-04
Identities = 36/127 (28%), Positives = 56/127 (43%), Gaps = 1/127 (0%)
Frame = +2
Query: 210 KFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLC 269
K G+K +SPDG+ L S+DG I V+D K L+ H PV
Sbjct: 536 KSGSKKFVLSVAWSPDGKRLACGSMDGTISVFDVQRAKFLHHLE--------GHFMPVRS 691
Query: 270 VDFS-RDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFD 328
+ +S D ++ S S DG + ++ + L H+ V V S +G+ + + S D
Sbjct: 692 LVYSPYDPRLLFSASDDGNVHMYDAE-GKALXGTMSXHASWVLCVDVSPNGAAIATGSSD 868
Query: 329 STARIHG 335
T R+ G
Sbjct: 869 KTVRLWG 889
>TC86503 similar to GP|15028127|gb|AAK76687.1 unknown protein {Arabidopsis
thaliana}, partial (54%)
Length = 1915
Score = 56.2 bits (134), Expect = 3e-08
Identities = 45/166 (27%), Positives = 77/166 (46%), Gaps = 8/166 (4%)
Frame = +3
Query: 229 LVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPV--LCVDFSRDSEMIASGSTDG 286
+++C D I+VWD +SG + TF H+ PV +C + + + I S + DG
Sbjct: 81 VITCGDDKTIKVWDAVSG--------AKQYTFEGHEAPVYSVCPHYKENIQFIFSTALDG 236
Query: 287 KIKVWRIRTAQCLRRLEH-AHSQGVTSVSFSRDGSQLLS--TSFDSTARI--HGLKSGKM 341
KIK W R+++ A + T++++S DG++L S TS D + I G +
Sbjct: 237 KIKAWLYDNLG--SRVDYDAPGRWCTTMAYSADGTRLFSCGTSKDGESSIVEWNESEGAV 410
Query: 342 LKEFRG-HTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQT 386
+ ++G + F R + A D ++K WD+ L T
Sbjct: 411 KRTYQGFRKRSMGVVQFDTTKNRFLAAGDDFSIKFWDMDNIQLLTT 548
Score = 37.0 bits (84), Expect = 0.018
Identities = 29/157 (18%), Positives = 70/157 (44%), Gaps = 1/157 (0%)
Frame = +1
Query: 264 DEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLL 323
++ V C S++ + S S GKI ++ + T + + +D + +
Sbjct: 1165 EDSVPCFALSKNDSYVMSAS-GGKISLFNMMTFKTMTTFMPPPPAATFLAFHPQDNNIIA 1341
Query: 324 STSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDC 383
DS+ +I+ ++ ++ + +GHT + F++ ++++ +D + VW+ +
Sbjct: 1342 IGMDDSSIQIYNVRVDEVKSKLKGHTKRITGLAFSHVLNVLVSSGADAQICVWNTDGWEK 1521
Query: 384 LQT-FKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKT 419
+T F PP R A ++ F ++ +V ++T
Sbjct: 1522 QKTRFLQLPPGRTPSAQSDTRVQFHQDQIQFLVVHET 1632
Score = 30.8 bits (68), Expect = 1.3
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 2/62 (3%)
Frame = +3
Query: 344 EFRGHTSYVNDATFTNDGTR--VITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVN 401
E H VND F++ + VIT D T+KVWD + TF+ G +A V
Sbjct: 15 EIDAHVCGVNDLAFSHPNKQLCVITCGDDKTIKVWDAVSGAKQYTFE------GHEAPVY 176
Query: 402 SV 403
SV
Sbjct: 177 SV 182
Score = 28.9 bits (63), Expect = 5.0
Identities = 41/190 (21%), Positives = 75/190 (38%), Gaps = 10/190 (5%)
Frame = +1
Query: 222 FSPDGQYLVSCSID-GFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIA 280
F P +++ +D I++++ ++K L+ H + + + FS ++
Sbjct: 1312 FHPQDNNIIAIGMDDSSIQIYNVRVDEVKSKLKG--------HTKRITGLAFSHVLNVLV 1467
Query: 281 SGSTDGKIKVWRIR--TAQCLRRLE-----HAHSQGVTSVSFSRDGSQLLSTSFDSTARI 333
S D +I VW Q R L+ +Q T V F +D Q L A
Sbjct: 1468 SSGADAQICVWNTDGWEKQKTRFLQLPPGRTPSAQSDTRVQFHQDQIQFLVVHETQLAIF 1647
Query: 334 HGLKSGKMLKEF--RGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPP 391
K + LK++ R ++ ++ ATF+ D + + D TV V++ PP
Sbjct: 1648 EATKL-ECLKQWAPRDSSAPISHATFSCDSQLIYASFLDATVCVFNASNLRLRCRINPPA 1824
Query: 392 PLRGGDASVN 401
L ++ N
Sbjct: 1825 YLPASVSNSN 1854
Score = 28.5 bits (62), Expect = 6.5
Identities = 37/177 (20%), Positives = 73/177 (40%), Gaps = 14/177 (7%)
Frame = +3
Query: 213 AKSHAECARFSPDGQYLVSCSIDGFIEVWDY----------ISGKLKKDLQYQAEETFMM 262
A ++ C + + Q++ S ++DG I+ W Y G+ + Y A+ T
Sbjct: 165 APVYSVCPHYKENIQFIFSTALDGKIKAWLYDNLGSRVDYDAPGRWCTTMAYSADGT--- 335
Query: 263 HDEPVLCVDFSRDSE--MIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGS 320
+ S+D E ++ ++G +K RT Q R+ S GV V F +
Sbjct: 336 ---RLFSCGTSKDGESSIVEWNESEGAVK----RTYQGFRK----RSMGV--VQFDTTKN 476
Query: 321 QLLSTSFDSTARIHGLKSGKMLK--EFRGHTSYVNDATFTNDGTRVITASSDCTVKV 375
+ L+ D + + + + ++L + G F DGT + +++D +K+
Sbjct: 477 RFLAAGDDFSIKFWDMDNIQLLTTVDADGGLPASPRIRFNKDGTLLAVSANDNGIKI 647
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,019,022
Number of Sequences: 36976
Number of extensions: 218775
Number of successful extensions: 1453
Number of sequences better than 10.0: 152
Number of HSP's better than 10.0 without gapping: 1156
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1361
length of query: 513
length of database: 9,014,727
effective HSP length: 100
effective length of query: 413
effective length of database: 5,317,127
effective search space: 2195973451
effective search space used: 2195973451
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142394.6


