

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.3 - phase: 0
(321 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
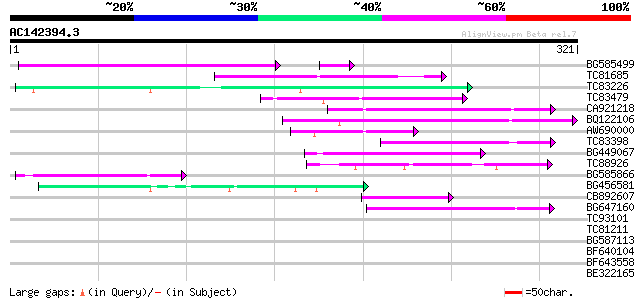
Score E
Sequences producing significant alignments: (bits) Value
BG585499 85 5e-19
TC81685 80 8e-16
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 63 2e-10
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 62 2e-10
CA921218 61 7e-10
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 56 2e-08
AW690000 53 1e-07
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 49 2e-06
BG449067 49 3e-06
TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {D... 47 8e-06
BG585866 47 8e-06
BG456581 45 3e-05
CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this... 43 1e-04
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 43 2e-04
TC93101 39 0.004
TC81211 38 0.005
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 38 0.006
BF640104 37 0.010
BF643558 34 0.066
BE322165 34 0.086
>BG585499
Length = 792
Score = 84.7 bits (208), Expect(2) = 5e-19
Identities = 46/150 (30%), Positives = 73/150 (48%), Gaps = 2/150 (1%)
Frame = +3
Query: 6 DNDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIGN-PMCRFCHDEIESE 64
D D W LW R P R + FM ++ HG + TN R+ + G C C + E+
Sbjct: 201 DQSIVDCDWKMLWGWRGPHRTQTFMWLVAHGCILTNYRRSRWGTRVLATCPCCGNADETV 380
Query: 65 IHVLRDCPKATALWLCVVDNAARTSFFE-GDLSSWIKFNLFTNVYWNNNVEWRDFWATAC 123
+HVL DC A+ +W+ +V + T+FF D W+ NL + +W+ + T C
Sbjct: 381 LHVLCDCRPASQVWIRLVPSDWITNFFSFDDCRDWVFKNLSKRSNGVSKFKWQPTFMTTC 560
Query: 124 HTIWNWRNKETHNDNYQRPLHAKNIIMIYV 153
+W WRNK + +QRP + +I ++
Sbjct: 561 WHMWTWRNKAIFEEGFQRPDNPTYVIQNFI 650
Score = 26.9 bits (58), Expect(2) = 5e-19
Identities = 9/20 (45%), Positives = 11/20 (55%)
Frame = +2
Query: 176 VGWKEPAIRWVKINTDGACK 195
+ WK P W K+N DG K
Sbjct: 719 IAWKRPLDGWAKLNCDGHAK 778
>TC81685
Length = 1706
Score = 80.5 bits (197), Expect = 8e-16
Identities = 47/131 (35%), Positives = 61/131 (45%)
Frame = -2
Query: 117 DFWATACHTIWNWRNKETHNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEEV 176
+ W+ CH+ WRNKE H + RP ++ YV Y + + + V
Sbjct: 352 ELWSITCHSFGMWRNKEEHCTGF*RPKKLWQQVINYVMRYKTPSYVNDNDVNAKQQIT-V 176
Query: 177 GWKEPAIRWVKINTDGACKDGSIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLE 236
W P W KIN+DG C+ AGCGGLI G GEWL GFSKF L+
Sbjct: 175 SWTLPQSDWDKINSDGMCQGSLRAGCGGLI*GDGGEWLCGFSKF--------------LK 38
Query: 237 GLRCAKRMGFT 247
LR +R GF+
Sbjct: 37 SLRLTQRYGFS 5
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 62.8 bits (151), Expect = 2e-10
Identities = 67/277 (24%), Positives = 109/277 (39%), Gaps = 18/277 (6%)
Frame = +3
Query: 4 NFDNDAQDE--IWLKLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIG-NPMCRFCHDE 60
N + + DE IW K+W++ R K + + + L K GI P+C CH +
Sbjct: 84 NNTSTSSDETLIWKKIWSLHTIPRHKVLLWRILNDSLPVRSSLRKRGIQCYPLCPRCHSK 263
Query: 61 IESEIHVLRDCPKATALW----LCV-VDNAARTSFFEGDLSSWI-KFNLFTNVYWNNNVE 114
E+ H+ CP + +W LC+ DN +F + + K T
Sbjct: 264 TETITHLFMSCPLSKRVWFGSNLCINFDNLPNPNFIN*LYEAIL*KDECITI*I------ 425
Query: 115 WRDFWATACHTIWNWRNKETHNDNYQRPLHAKNIIMIYVNDYHNAIAKF--------VIV 166
A + +W+ RN D + ++DY A +
Sbjct: 426 -----AAIIYNLWHARNLSVLEDQTILEMDIIQRASNCISDYKQANTQAPPSMARTGYDP 590
Query: 167 TSQPRHLEEVGWKEPAIRWVKINTDGACKDGSIAGCGGLIRGSEGEWLAGFS-KFLGKCD 225
SQ R + WK P + VK+NTD ++ G G +IR G +A + + G
Sbjct: 591 RSQHRPAKNTKWKRPNLGLVKVNTDANLQNHGKWGLGIIIRDEVGLVMAASTWETDGNDR 770
Query: 226 AFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNII 262
A AE + +L G+R AK GF V D+ ++ ++
Sbjct: 771 ALEAEAYALLTGMRFAKDCGFXKVXFEGDNEKLMKMV 881
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein
{Plasmodium falciparum 3D7}, partial (0%)
Length = 1222
Score = 62.4 bits (150), Expect = 2e-10
Identities = 40/128 (31%), Positives = 61/128 (47%), Gaps = 11/128 (8%)
Frame = +2
Query: 143 LHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEEV-----------GWKEPAIRWVKINTD 191
LHAKNI YV + K +++ + + WK+P I W K+NTD
Sbjct: 845 LHAKNIP--YVLSFTQERIKGMVLLKNLNQIANILNPVSRSIIWCEWKKPEIGWTKLNTD 1018
Query: 192 GACKDGSIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVEL 251
G+ + AG GGL+R GE + F + D F+ ELW + GL + +G ++ +
Sbjct: 1019GSVNKET-AGFGGLLRDYRGEPICAFVSKAPQGDTFLVELWAIWRGLVLSLGLGIKSIWV 1195
Query: 252 NVDSLVVV 259
DS+ VV
Sbjct: 1196ESDSMSVV 1219
>CA921218
Length = 707
Score = 60.8 bits (146), Expect = 7e-10
Identities = 40/129 (31%), Positives = 63/129 (48%)
Frame = -3
Query: 181 PAIRWVKINTDGACKDGSIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRC 240
P W+ NT+G+ + A CGG R S +L F++ G +A A+L G + +
Sbjct: 594 PEPLWINCNTNGSANINTSA-CGGTFRNSNAYFLLCFAENTGNGNALHAKLSGAMRAIEI 418
Query: 241 AKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANR 300
A ++ + L +DS +VVN S+ + + R+ L+ V H +RE N+
Sbjct: 417 AAARNWSHLWLELDSSLVVNAFKSKSVIHWNLRNRWNNCLFLIS-SMNFFVSHVFREGNQ 241
Query: 301 CADALANIG 309
CAD LAN G
Sbjct: 240 CADGLANFG 214
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 55.8 bits (133), Expect = 2e-08
Identities = 47/177 (26%), Positives = 76/177 (42%), Gaps = 10/177 (5%)
Frame = -2
Query: 155 DYHNAIAKFVIVTSQPRHLEEVGWK-EPAIRW-------VKINTDGACKDGS-IAGCGGL 205
D H+ +++T+QP + + ++W +N DG+C G GL
Sbjct: 662 DLHSIFKTLLMLTAQPFLYNSLSQATDTVVKWNCDNNSCYILNVDGSCLGNP*PTGFNGL 483
Query: 206 IRGSEGEWLAGF-SKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITS 264
IR G + +GF D +AEL + +GLR MG + DSL V++I
Sbjct: 482 IRNIAGLFNSGFPGNITNTSDILLAELHAIFQGLRMISDMGISDFVCYFDSLHYVSLING 303
Query: 265 ERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALANIGCIMGNEMMFYES 321
+L+Q I+ L+ + K V H+ E N CAD L +G + + + S
Sbjct: 302 PSMKFHVYATLIQDIKDLV-ITSKASVFHTLCEGNYCADFLEMLGAASDSVLTIHVS 135
>AW690000
Length = 652
Score = 53.1 bits (126), Expect = 1e-07
Identities = 28/74 (37%), Positives = 43/74 (57%), Gaps = 2/74 (2%)
Frame = +3
Query: 160 IAKFVIVTSQPR--HLEEVGWKEPAIRWVKINTDGACKDGSIAGCGGLIRGSEGEWLAGF 217
+ KF + P+ + EV W+ P W+K NTDG+ + S A CGG+ R + + L F
Sbjct: 351 LKKFNVTIHPPKAPKIIEVIWRPPIPHWIKCNTDGSSRSHSSA-CGGIFRNHDTDLLLCF 527
Query: 218 SKFLGKCDAFIAEL 231
++ G+C+AF AEL
Sbjct: 528 AENTGECNAFHAEL 569
Score = 30.0 bits (66), Expect = 1.2
Identities = 13/46 (28%), Positives = 19/46 (41%)
Frame = +2
Query: 53 MCRFCHDEIESEIHVLRDCPKATALWLCVVDNAARTSFFEGDLSSW 98
MC C + ES +H+ +C A LW +T F+ W
Sbjct: 11 MCSLCCKQAESSLHLFFECSYAVNLWCWFASVLNKTLHFQSLEDMW 148
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 49.3 bits (116), Expect = 2e-06
Identities = 34/100 (34%), Positives = 51/100 (51%), Gaps = 1/100 (1%)
Frame = +2
Query: 211 GEWLAGFSKFLGKC-DAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESN 269
G + +GFS + D AEL +L GL+ A+ + V DSL VN+I
Sbjct: 89 GFFNSGFSGHIDHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLINGPSVVY 268
Query: 270 ASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALANIG 309
+ +L+Q I+ L+ ++ H+ RE NRCAD LA +G
Sbjct: 269 HAYATLIQDIKDLI----RLSKLHTLREGNRCADFLAKLG 376
>BG449067
Length = 578
Score = 48.9 bits (115), Expect = 3e-06
Identities = 30/103 (29%), Positives = 50/103 (48%), Gaps = 1/103 (0%)
Frame = -2
Query: 168 SQPRHLEEVGWKEPAIRWVKINTDGACKDGSIAGCGGLIRGSEGEWLAGFSKF-LGKCDA 226
S P+H++ WK+P +K+N+D + G G + R EG +A + F G A
Sbjct: 328 SVPKHVK---WKKPEKDIIKLNSDANLSSTDLWGIGVVARNDEGFAMASGTWFRFGFPSA 158
Query: 227 FIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESN 269
AE WG+ + + A GF+ V+ D+ V+ ++ E N
Sbjct: 157 TTAEAWGIYQAMIFAGEYGFSKVQFESDNERVIQMLNGTEEVN 29
>TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {Drosophila
melanogaster}, partial (1%)
Length = 1073
Score = 47.4 bits (111), Expect = 8e-06
Identities = 47/149 (31%), Positives = 68/149 (45%), Gaps = 10/149 (6%)
Frame = +3
Query: 169 QPRHLEEVGWKEPAIRWVKINTDGAC---KDGSIAGCGGLIRGSEGEWLAGFSKFLG--- 222
+P +LEEV +N DG+ ++ AGCGG++ S G+WL GF++ L
Sbjct: 321 RPDYLEEV----------ILNVDGSLLREREVPSAGCGGVLSDSSGKWLCGFAQKLNPNL 470
Query: 223 KCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRS---LVQKI 279
K D E +L GL K G + + D+ VV + N GRS LV I
Sbjct: 471 KVDE--TEKEAILRGLLWVKEKGKRKILVKSDNEGVVYSV------NCGGRSNDPLVCGI 626
Query: 280 RKLLQM-EWKVKVKHSYREANRCADALAN 307
R LL W+ + + +N AD LA+
Sbjct: 627 RDLLNSPHWEATLTCIHGRSNAVADRLAH 713
>BG585866
Length = 828
Score = 47.4 bits (111), Expect = 8e-06
Identities = 24/98 (24%), Positives = 48/98 (48%), Gaps = 1/98 (1%)
Frame = +3
Query: 4 NFDNDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLST-NLRKHKMGIGNPMCRFCHDEIE 62
N++N + W +W ++ PE+ K F+ + H + T +L H+ + + +C C + E
Sbjct: 435 NYNNSS----WSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEE 602
Query: 63 SEIHVLRDCPKATALWLCVVDNAARTSFFEGDLSSWIK 100
S H +RDC + +W + ++ F + W+K
Sbjct: 603 SFFHCVRDCRFSKIIW-HKIGFSSPDFFSSSSVQDWLK 713
>BG456581
Length = 683
Score = 45.4 bits (106), Expect = 3e-05
Identities = 53/209 (25%), Positives = 81/209 (38%), Gaps = 22/209 (10%)
Frame = +2
Query: 17 LWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIGN-PMCRFCHDEIESEIHVLRDCPKAT 75
+WN P L H RL T+ G MC FC +++E+ H+ C
Sbjct: 80 IWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCALVSMCSFCLEQVETSDHLFLRCKFVV 259
Query: 76 ALW--LCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATAC----HTIWNW 129
LW LC + R D SS+ L +++ + + + RD + A H+IW W
Sbjct: 260 TLWSWLC---SQLRVGL---DFSSFKA--LLSSLPRHCSSQVRDLYVAAVVHMVHSIW-W 412
Query: 130 RNKETHNDNYQRPLHAKNIIMIYVNDYHNAI---------AKFVIVTSQPRH------LE 174
+ + HA + + + A+ A + V P H +
Sbjct: 413 ARNNVRFSSAKVSAHAVQVRVHALIGLSGAVSTGKCIAADAAILDVFRIPPHRRSMXEMV 592
Query: 175 EVGWKEPAIRWVKINTDGACKDGSIAGCG 203
V WK P+ WVK NTDG+ + S A G
Sbjct: 593 SVCWKPPSAPWVKGNTDGSXLNNSGAXGG 679
>CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (1%)
Length = 782
Score = 43.1 bits (100), Expect = 1e-04
Identities = 18/52 (34%), Positives = 29/52 (55%)
Frame = +1
Query: 200 AGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVEL 251
A CGGLI+ +G ++ ++ LG C AELWG+ GL G++ + +
Sbjct: 622 AACGGLIQDDQGHFVFHYANKLGSCSVLQAELWGI*HGLSIDWNRGYSKIRV 777
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides},
partial (2%)
Length = 780
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/107 (26%), Positives = 51/107 (47%), Gaps = 1/107 (0%)
Frame = +1
Query: 203 GGLIRGSEGEWLAGFSKFLGKC-DAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNI 261
GG+IR ++ GFS F+ + AEL + + L+ A + + DSL+ VN+
Sbjct: 379 GGVIRNYLSTYITGFSGFISIYQNIMFAELTTLHQSLKLAISLNIEEMVCYSDSLLTVNL 558
Query: 262 ITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALANI 308
I + + L+ I+ ++ + H+ RE N+CAD + +
Sbjct: 559 IKEDISQHHVYAVLIHNIKYIMSSR-NFTLHHTLREGNQCADFMVKL 696
>TC93101
Length = 675
Score = 38.5 bits (88), Expect = 0.004
Identities = 48/229 (20%), Positives = 87/229 (37%), Gaps = 16/229 (6%)
Frame = -1
Query: 34 YHGRLSTNLRKHKMGIG-NPMCRFCHDEIESEIHVLRDCPKATALWLCVVDNAARTSFFE 92
+H L +K G+ P+C C+ +E+ H+ C + +W +
Sbjct: 675 FHTTLPVRXELNKRGVNCPPLCPRCYFNLETTNHIFMSCERTQRVWFGSQLSIRFPDNST 496
Query: 93 GDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKETHNDNYQRPLH----AKNI 148
+ S W+ F+ +N ++ + ++IW+ RNK + + A+N
Sbjct: 495 INFSDWL-FDAISNQTEEIIIKI----SAITYSIWHARNKAIFENQFVSEDTIIQ*AQNS 331
Query: 149 IMIYVN-----DYHNAIAKFVIVTSQP-----RHLEEVGWKEPAIRWVKINTDGACKDGS 198
I+ Y N + + TS R W++P +K N D +
Sbjct: 330 ILAYEQATIKPQNPNIVLSSLSATSNTNTTRRRSNVRSRWQKPLNNILKANCDANLQVQG 151
Query: 199 IAGCGGLIRGSEGEWLAGFSKFLGKCD-AFIAELWGVLEGLRCAKRMGF 246
G G +IR ++GE + + D A AE + +L + AK GF
Sbjct: 150 RWGLGCIIRNADGEAKVTATWCINGFDCAATAETYAILAAMYLAKDYGF 4
>TC81211
Length = 509
Score = 38.1 bits (87), Expect = 0.005
Identities = 15/25 (60%), Positives = 21/25 (84%)
Frame = +1
Query: 285 MEWKVKVKHSYREANRCADALANIG 309
++W++K+ HSYRE NR ADALA +G
Sbjct: 379 LDWEIKICHSYREFNRFADALAYMG 453
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 37.7 bits (86), Expect = 0.006
Identities = 19/75 (25%), Positives = 35/75 (46%), Gaps = 2/75 (2%)
Frame = -3
Query: 6 DNDAQDEIWLKLWNIRAPERIKHFMSMLYHGRLST--NLRKHKMGIGNPMCRFCHDEIES 63
D + D+++ ++W +++HF+ L T N+R + + C C E E+
Sbjct: 321 DQPSLDDLYQRVWKYNTSPKVRHFLWRCISNSLPTAANMRSRHIS-KDGSCSRCGMESET 145
Query: 64 EIHVLRDCPKATALW 78
H+L CP A +W
Sbjct: 144 VNHILFQCPYARLIW 100
>BF640104
Length = 344
Score = 37.0 bits (84), Expect = 0.010
Identities = 31/110 (28%), Positives = 48/110 (43%), Gaps = 2/110 (1%)
Frame = -1
Query: 176 VGWKEPAIRWVKINTDGACKDGSIAGCGGLIRGSEGEWLA-GFSKFLGKCDAFIAELWGV 234
V W +P +KIN D + G G + R G +A G G AE WGV
Sbjct: 344 VKWIKPHQGVIKINCDANLTSEDVWGIGVITRNDNGIVMASGTWNRPGFMCPITAEAWGV 165
Query: 235 LEGLRCAKRMGFTAVELNVDSLVVVNIITSERESNASG-RSLVQKIRKLL 283
+ A GF V D+ ++++++ E E + S S++Q I L+
Sbjct: 164 YQAALFALDQGFQNVLFENDNEKLISMLSREEEGHRSYLGSIIQSIISLV 15
>BF643558
Length = 645
Score = 34.3 bits (77), Expect = 0.066
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Frame = +2
Query: 160 IAKFVIVTSQPR--HLEEVGWKEPAIRWVKINTDGACKDGSIAGCGGLIR 207
+ KF I P+ + EV W P + W+K NTDG+ + + + CGG I+
Sbjct: 497 LKKFSIDIHPPKAPKIIEVLWNPPPLHWIKCNTDGS-SNTNTSSCGGDIQ 643
>BE322165
Length = 650
Score = 33.9 bits (76), Expect = 0.086
Identities = 21/70 (30%), Positives = 33/70 (47%), Gaps = 1/70 (1%)
Frame = -3
Query: 201 GCGGLIRGSEGEWLAGFSKF-LGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVV 259
G G + R EG +A + F G A AE WG+ + + A GF+ V+ D+ V+
Sbjct: 246 GIGVVARNDEGFAMASGTWFRFGFPSATTAEAWGIYQAMIFAGEYGFSKVQFESDNERVI 67
Query: 260 NIITSERESN 269
++ E N
Sbjct: 66 QMLNGTEEVN 37
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.450
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,344,318
Number of Sequences: 36976
Number of extensions: 228783
Number of successful extensions: 1158
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1135
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1147
length of query: 321
length of database: 9,014,727
effective HSP length: 96
effective length of query: 225
effective length of database: 5,465,031
effective search space: 1229631975
effective search space used: 1229631975
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC142394.3


