

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.1 - phase: 0 /pseudo
(426 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
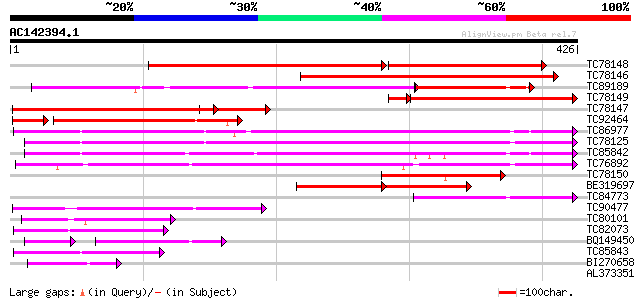
Score E
Sequences producing significant alignments: (bits) Value
TC78148 weakly similar to GP|4508070|gb|AAD21414.1| 24349 {Arabi... 289 e-132
TC78146 weakly similar to GP|4508070|gb|AAD21414.1| 24349 {Arabi... 335 3e-92
TC89189 weakly similar to GP|6469618|gb|AAF13357.1| (3R)-linaloo... 207 3e-78
TC78149 weakly similar to GP|4508070|gb|AAD21414.1| 24349 {Arabi... 223 2e-61
TC78147 226 2e-59
TC92464 weakly similar to GP|23495894|dbj|BAC20102. sesquiterpen... 169 8e-49
TC86977 weakly similar to GP|19880625|gb|AAM00426.1 putative ter... 175 3e-44
TC78125 weakly similar to GP|16751271|gb|AAL25826.1 cyclase {Nic... 173 1e-43
TC85842 weakly similar to GP|5360741|dbj|BAA82141.1 vetispiradie... 168 3e-42
TC76892 weakly similar to GP|9864189|gb|AAG01339.1| terpene synt... 167 5e-42
TC78150 147 6e-36
BE319697 weakly similar to GP|4508070|gb|A 24349 {Arabidopsis th... 119 2e-27
TC84773 weakly similar to GP|17227146|gb|AAL38029.1 linalool syn... 96 3e-20
TC90477 weakly similar to GP|9864189|gb|AAG01339.1| terpene synt... 75 5e-14
TC80101 weakly similar to GP|9864189|gb|AAG01339.1| terpene synt... 73 2e-13
TC82073 weakly similar to GP|14331015|emb|CAC41012. putative chl... 70 1e-12
BQ149450 weakly similar to GP|5360741|dbj vetispiradiene synthas... 54 2e-11
TC85843 weakly similar to GP|3687297|emb|CAA06614.1 5-epi-aristo... 56 3e-08
BI270658 weakly similar to PIR|T09672|T096 ent-kaurene synthase ... 43 2e-04
AL373351 weakly similar to PIR|T52059|T520 ent-kaurene synthase ... 41 0.001
>TC78148 weakly similar to GP|4508070|gb|AAD21414.1| 24349 {Arabidopsis
thaliana}, partial (16%)
Length = 902
Score = 289 bits (739), Expect(2) = e-132
Identities = 131/179 (73%), Positives = 161/179 (89%)
Frame = +3
Query: 105 LIALYEASQLSIEGEDGLNDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRF 164
LIALYEASQLSIEGED LND+GYL ELL AWL R+Q+H +AI+V+NT+Q P+HYGLSRF
Sbjct: 3 LIALYEASQLSIEGEDSLNDVGYLCSELLHAWLSRNQEHNEAIHVANTVQNPLHYGLSRF 182
Query: 165 MDKSIFINDLKAKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYN 224
MDKS FI+DLKA+ +CL+ELAK+NS+IV+FMN+NE+VEV KWW++LGL KE+KF+GY
Sbjct: 183 MDKSTFIHDLKAEKDLMCLEELAKINSTIVRFMNQNETVEVSKWWKELGLDKEVKFSGYQ 362
Query: 225 PLKWYMWPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEM 283
PLKWY WPMACFTDP FS +R+ELTKPISL+Y+IDDIFDVH TL+QLT+FT+A+NRWE+
Sbjct: 363 PLKWYTWPMACFTDPNFSEQRIELTKPISLIYVIDDIFDVHATLEQLTIFTDAINRWEI 539
Score = 201 bits (510), Expect(2) = e-132
Identities = 93/119 (78%), Positives = 107/119 (89%)
Frame = +1
Query: 285 GAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSG 344
G E LPNFMK+SL++LY++TNNFAE VYKKHGFNPIDTLK SW+RLLNAF++EAHWLNSG
Sbjct: 544 GTEQLPNFMKISLNALYEITNNFAEKVYKKHGFNPIDTLKKSWIRLLNAFMEEAHWLNSG 723
Query: 345 ILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILR 403
LP E+YLNNGIVSTGVHVVL HAFFL+DH GITK+T+ ILDE+F N+IYSVAKILR
Sbjct: 724 HLPKAEDYLNNGIVSTGVHVVLEHAFFLLDHVNGITKQTIDILDEKFXNVIYSVAKILR 900
>TC78146 weakly similar to GP|4508070|gb|AAD21414.1| 24349 {Arabidopsis
thaliana}, partial (13%)
Length = 1010
Score = 335 bits (858), Expect = 3e-92
Identities = 153/194 (78%), Positives = 174/194 (88%)
Frame = +3
Query: 219 KFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAV 278
KF+ Y PLKWY WPMACFTDP FS +R+ELTKPISL+Y+IDDIFDVH TLDQLT+FT+AV
Sbjct: 3 KFSEYQPLKWYTWPMACFTDPNFSEQRIELTKPISLIYVIDDIFDVHATLDQLTIFTDAV 182
Query: 279 NRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEA 338
NRWE G E LPNFMK++L++LY +TN+FAEMVYKKHGFNPIDTLK SW+ LLNAF++EA
Sbjct: 183 NRWEFTGTEQLPNFMKIALNALYDITNSFAEMVYKKHGFNPIDTLKKSWILLLNAFMEEA 362
Query: 339 HWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSV 398
HWLNSG LP E+YLNNGIVSTGVHVVLIHAFFL+DH GITKET+ ILDE+FPN+IYSV
Sbjct: 363 HWLNSGHLPRAEDYLNNGIVSTGVHVVLIHAFFLLDHVNGITKETIDILDEKFPNVIYSV 542
Query: 399 AKILRLSDDLEGVK 412
AKILRLSDDLEG K
Sbjct: 543 AKILRLSDDLEGAK 584
Score = 34.7 bits (78), Expect = 0.072
Identities = 16/28 (57%), Positives = 22/28 (78%), Gaps = 1/28 (3%)
Frame = +1
Query: 400 KILRLSDDLEG-VKSGDQNGLDGSYLNC 426
K+L + + G ++SGDQNGLDGSYL+C
Sbjct: 664 KVLIIRTIIHG*MQSGDQNGLDGSYLDC 747
>TC89189 weakly similar to GP|6469618|gb|AAF13357.1| (3R)-linalool synthase
{Artemisia annua}, partial (11%)
Length = 1409
Score = 207 bits (526), Expect(2) = 3e-78
Identities = 118/298 (39%), Positives = 171/298 (56%), Gaps = 6/298 (2%)
Frame = +2
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E ++D IQRLG+E+ F EEI+ LE++ +L L + A FR+LRQ G
Sbjct: 251 ECLSMVDSIQRLGMEYNFEEEIEATLERKHTMLRFQNFQRNEYQGLSQAAFQFRMLRQEG 430
Query: 77 YYVNAELFDCLKCSKKS------LRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSC 130
YY+ ++FD +++ LR+ E + L L A + I L+++G
Sbjct: 431 YYITPDIFDKFCDNQRQTSSIHFLRI*M-E*LHCLKLLN*A*KEKI----CLDNVGQFCG 595
Query: 131 ELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMN 190
+ L W H QA +V++TL YP H LSRF + + N + + +K++
Sbjct: 596 QYLNDWSSTFHGHPQAKFVAHTLMYPTHKTLSRFTPTIMQSQNATWTNS---IQQFSKID 766
Query: 191 SSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTK 250
+ +V + E V KWW+DLGL K+++FA P+KWY W MAC DP FS ER+ELTK
Sbjct: 767 TQMVSSSHLKEIFAVSKWWKDLGLPKDLEFARDEPIKWYSWSMACLPDPQFSEERIELTK 946
Query: 251 PISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFA 308
P+SL+YIIDDIFD +G +D+LTLFT+AV RW++ E LP+ MKV +LY +TN FA
Sbjct: 947 PLSLIYIIDDIFDFYGNIDELTLFTDAVKRWDLSAIEQLPDCMKVCFKALYDITNEFA 1120
Score = 103 bits (257), Expect(2) = 3e-78
Identities = 51/89 (57%), Positives = 67/89 (74%)
Frame = +3
Query: 306 NFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVV 365
N Y KHG+NP+ +L SWVRLLNAFL+EA W SG +P +EEYL N IVSTGVHV+
Sbjct: 1113 NLLLRTYIKHGWNPLTSLIKSWVRLLNAFLQEAKWFASGNVPKSEEYLKNAIVSTGVHVI 1292
Query: 366 LIHAFFLMDHAKGITKETLSILDEEFPNI 394
L+HAFF M +GIT++T+S++D +FP +
Sbjct: 1293 LVHAFFCM--GQGITEKTVSLMD-DFPTL 1370
>TC78149 weakly similar to GP|4508070|gb|AAD21414.1| 24349 {Arabidopsis
thaliana}, partial (10%)
Length = 864
Score = 223 bits (567), Expect(2) = 2e-61
Identities = 102/125 (81%), Positives = 117/125 (93%)
Frame = +1
Query: 302 KVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTG 361
++T+NFA+MVYKKHGFNPIDTLK SW+RLLNAF++EAHWLNSG LP E+YLNNGIVSTG
Sbjct: 70 EITSNFADMVYKKHGFNPIDTLKKSWIRLLNAFMEEAHWLNSGHLPRAEDYLNNGIVSTG 249
Query: 362 VHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDG 421
VHVVL+HAFFL+DH GITKET+ ILDE+FPN+IYSVAKILRLSDDLEG KSG+QNGLDG
Sbjct: 250 VHVVLVHAFFLLDHVHGITKETIDILDEKFPNVIYSVAKILRLSDDLEGAKSGEQNGLDG 429
Query: 422 SYLNC 426
SYL+C
Sbjct: 430 SYLDC 444
Score = 31.6 bits (70), Expect(2) = 2e-61
Identities = 12/18 (66%), Positives = 16/18 (88%)
Frame = +3
Query: 285 GAENLPNFMKVSLSSLYK 302
G E LPNFMK+SL++LY+
Sbjct: 18 GTEQLPNFMKISLNALYR 71
>TC78147
Length = 662
Score = 226 bits (575), Expect = 2e-59
Identities = 112/155 (72%), Positives = 132/155 (84%)
Frame = +1
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
KQV+KKLV ED E+F+LIDIIQR GIEHYF EEIKVALEK LIL++NPIDFV+SHEL
Sbjct: 79 KQVYKKLVKIEDSMESFYLIDIIQRFGIEHYFAEEIKVALEKLHLILNTNPIDFVNSHEL 258
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
YE +LAFRLLRQGG+YVNA+LFD LKC+K+ K+GED+KGLIALYEASQLSI+GED L
Sbjct: 259 YEFSLAFRLLRQGGHYVNADLFDSLKCNKRMFEEKHGEDLKGLIALYEASQLSIDGEDSL 438
Query: 123 NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPI 157
ND+GYL ELL WL R+Q+H +AI+V NTLQ P+
Sbjct: 439 NDVGYLCRELLHNWLSRNQEHNEAIHVVNTLQNPL 543
Score = 63.9 bits (154), Expect = 1e-10
Identities = 33/55 (60%), Positives = 43/55 (78%), Gaps = 1/55 (1%)
Frame = +2
Query: 143 HIQAIYVSNTL-QYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVKF 196
+I +Y+ TL + HYGLSRFMDKS FI+DLKA+ ICL+ELAK+NS+IV+F
Sbjct: 497 NIMKLYML*TLFRILFHYGLSRFMDKSTFIHDLKAEKDLICLEELAKINSTIVRF 661
>TC92464 weakly similar to GP|23495894|dbj|BAC20102. sesquiterpene
synthase-like protein {Oryza sativa (japonica
cultivar-group)}, partial (4%)
Length = 753
Score = 169 bits (427), Expect(2) = 8e-49
Identities = 93/145 (64%), Positives = 111/145 (76%), Gaps = 3/145 (2%)
Frame = +3
Query: 34 FVEEIKVALEKQFLILSSNPI-DFVSSHELYEVALAFRLLRQGGYYVNAELFDCLKCSKK 92
F EEIKVALEK LIL++NPI DFVSSHELYEVALAFRLLRQGG+YVN +LFD LKC+K+
Sbjct: 315 FAEEIKVALEKLHLILNTNPIIDFVSSHELYEVALAFRLLRQGGHYVNPDLFDNLKCTKR 494
Query: 93 SLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAWLPRHQDHIQAIYVSNT 152
K+GEDVKGLIALYEASQLSIEGED LND+GYL CELL AWL + + ++
Sbjct: 495 MFEEKHGEDVKGLIALYEASQLSIEGEDCLNDVGYLCCELLHAWLSK-KPRT*GCFICCK 671
Query: 153 LQYPIHYGLS--RFMDKSIFINDLK 175
IH+ ++ RF+DK+ +DLK
Sbjct: 672 HSSRIHFTMACQRFLDKAHSNHDLK 746
Score = 43.1 bits (100), Expect(2) = 8e-49
Identities = 20/27 (74%), Positives = 23/27 (85%)
Frame = +1
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLG 29
KQV+KKLV EDP E+F+LIDIIQR G
Sbjct: 220 KQVYKKLVKIEDPMESFYLIDIIQRFG 300
>TC86977 weakly similar to GP|19880625|gb|AAM00426.1 putative terpene
synthase {Citrus x paradisi}, partial (49%)
Length = 1812
Score = 175 bits (444), Expect = 3e-44
Identities = 117/427 (27%), Positives = 214/427 (49%), Gaps = 4/427 (0%)
Frame = +2
Query: 4 QVHKKLVT-SEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
+V LV+ +E P LID I RLG+ ++F +EI L+ N + L
Sbjct: 218 EVRNMLVSKTEMPLTKVKLIDSICRLGVGYHFEKEIDEVLQHIHKSYVENG-EITLEDNL 394
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
+A+ FR+ RQ G +V+ +F+ K + + DV+G+++LYEAS + + +D L
Sbjct: 395 CSLAMLFRVFRQQGLHVSPNVFNKFKDKQGNFNENLSTDVEGMLSLYEASHMMVHEDDIL 574
Query: 123 NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDK---SIFINDLKAKNK 179
+ + L++ + + A + TL+ +H + R + SI+ D
Sbjct: 575 EEALNFTSTHLESIASQSSPSLAA-QIEYTLKQALHKNIPRLEARHYISIYEKDPTCDE- 748
Query: 180 WICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDP 239
L AK++ ++++ +++ E + KWW++L + ++ +A + W + F +P
Sbjct: 749 --VLLTFAKLDFNLLQSLHQKEFGNISKWWKELDFSTKLPYARDRIAECSFWVLTAFFEP 922
Query: 240 CFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSS 299
+S R + K I+L+ IIDD +D +GT+D+L LFT+AV RW++ + LP++MK S
Sbjct: 923 QYSQARKMMIKVITLLSIIDDTYDAYGTIDELELFTKAVERWDISSLDELPDYMKPIYRS 1102
Query: 300 LYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVS 359
+ + + K+ +D KI + + + AF+ EA WLN +PTTEEY+ S
Sbjct: 1103FLTIYEEIEKEMRKEGRIYTLDYYKIEFKKSVQAFMTEARWLNENHIPTTEEYMRISKKS 1282
Query: 360 TGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGL 419
++++ ++ M TKE + + E P I+ + A + RL D++ + + G
Sbjct: 1283GAYPLLILTSYIGMGDI--ATKEIFNWVSNE-PRIVNAAATLCRLMDEIVSSEFEQKRGH 1453
Query: 420 DGSYLNC 426
S L+C
Sbjct: 1454VCSLLDC 1474
>TC78125 weakly similar to GP|16751271|gb|AAL25826.1 cyclase {Nicotiana
tabacum}, partial (40%)
Length = 1954
Score = 173 bits (439), Expect = 1e-43
Identities = 108/415 (26%), Positives = 214/415 (51%)
Frame = +2
Query: 12 SEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRL 71
+E P+E +LID I RLG+ +++ EI ++ N + L +++ FRL
Sbjct: 218 TEKPSEKINLIDSICRLGVSYHYENEIDEVMQYIHKSYVENG-EITLQDNLCTLSVLFRL 394
Query: 72 LRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCE 131
LRQ G++V+ +F+ K + + + DV+G+++LYEAS + + GE+ L +
Sbjct: 395 LRQQGFHVSPNVFNQFKDEQGNFSERLITDVEGMLSLYEASHMMVHGEEILEKALAFTST 574
Query: 132 LLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNS 191
L++ + + + A V ++L+ +H R + K + L LAK++
Sbjct: 575 HLESIVTQLSPFLAA-QVKHSLKQALHRNFHRLEARRYISIYEKDPSHNETLLTLAKLDF 751
Query: 192 SIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKP 251
+I++ +++ E ++ KWW+++ + K + + ++ W +A + +P +S R++LTK
Sbjct: 752 NILQCLHRKEFGKICKWWKEIDIPKNLPYVRDRIVEMCFWVLAVYFEPQYSQARIKLTKL 931
Query: 252 ISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMV 311
+L+ IIDD +D +G++D+L LFT+A+ RW++ + LPN+MK+ S+ KV E +
Sbjct: 932 GALLSIIDDTYDAYGSIDELELFTKAIERWDISSMDGLPNYMKLIYISVLKVLEEVEEDM 1111
Query: 312 YKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFF 371
K+ + + ++ A++ EA W N+ +PT EEYL ++ ++ ++
Sbjct: 1112TKEGRLYTLKYYIKEFQMVVQAYVTEARWFNNNYVPTVEEYLQISKITCCHSLLTTSSYI 1291
Query: 372 LMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
M E + +P I+ +++ I RL D++ + + G S L+C
Sbjct: 1292GMGET---ATENIFKWVSNWPKIVDAISTICRLMDEIVTSEFERKRGHVCSLLDC 1447
>TC85842 weakly similar to GP|5360741|dbj|BAA82141.1 vetispiradiene synthase
{Solanum tuberosum}, partial (38%)
Length = 1955
Score = 168 bits (426), Expect = 3e-42
Identities = 119/448 (26%), Positives = 219/448 (48%), Gaps = 33/448 (7%)
Frame = +1
Query: 12 SEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRL 71
SE P E LID I RLG+ ++F +EI L+ N + + L+ +A+ FR+
Sbjct: 238 SEKPFEKVKLIDSICRLGLSYHFEKEIDEVLQHIHKSYVENG-EIILEDNLFSLAVLFRV 414
Query: 72 LRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND-LGYLSC 130
LRQ G+YV+ +F K + + DV+G+++LYEAS L + GED L + L + S
Sbjct: 415 LRQHGFYVSPNVFTKFKDEQGNFNETLIMDVEGMLSLYEASHLIVHGEDILEEALAFTST 594
Query: 131 ELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKS-IFINDLKAKNKWICLDELAKM 189
L ++ H A V L+ +H L R + I I + + I L +K+
Sbjct: 595 HL--EFIATESSHSLAAQVKYALRQALHKSLPRLEARRYISIYEQDPSHDEILLT-FSKL 765
Query: 190 NSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELT 249
+ ++++ +++ E + KWW++L + ++ +A ++ W + + +P +S R L
Sbjct: 766 DFNLLQSLHQKEFGNISKWWKELDFSSKLPYARDRIVECCFWTLTVYFEPQYSRARKMLP 945
Query: 250 KPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKV------ 303
K ++ +IDD +D +GT+D+L FTEA+ RW++ ++++P++MK+ S + V
Sbjct: 946 KINVMLSLIDDTYDSYGTIDELERFTEAIERWDVIVSDDIPDYMKLLYKSFWNVYEEIEQ 1125
Query: 304 ----------TNNFAEMVYKK--HGFNPIDTLKI-------------SWVRLLNAFLKEA 338
N + +++YK + + I+ I + + + A++ EA
Sbjct: 1126AVIEEGREYILNYYKKLLYKSFWNVYEEIEQAMIEEGREYILNYYKKEFKKAVQAYMTEA 1305
Query: 339 HWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSV 398
WLN +PTTEEY+ S ++++ ++ M +T+ + E P I+
Sbjct: 1306RWLNENYIPTTEEYMRVSRTSCCYSLLILASYIGM--GDKVTENIFKWVTNE-PKIVNGA 1476
Query: 399 AKILRLSDDLEGVKSGDQNGLDGSYLNC 426
A I RL D++ + + G S L+C
Sbjct: 1477ANICRLMDEIVSTEFEQKRGHVCSLLDC 1560
>TC76892 weakly similar to GP|9864189|gb|AAG01339.1| terpene synthase
{Citrus junos}, partial (37%)
Length = 2245
Score = 167 bits (424), Expect = 5e-42
Identities = 114/430 (26%), Positives = 211/430 (48%), Gaps = 8/430 (1%)
Frame = +2
Query: 5 VHKKLVTSEDPT-ENFHLIDIIQRLGIEHYF---VEEIKVALEKQFLILSSNPIDFVSSH 60
V LV++ + T HLID I RLG+ ++F +EE+ + K +++ ++ H
Sbjct: 173 VRNMLVSNTEKTLAKIHLIDSIYRLGVSYHFEQEIEEVLHGIHKNYVVNGEITLE---DH 343
Query: 61 ELYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGED 120
L +A+ FRLLRQ G +V +F+ K + K ++V+G+++LYEA+ + + GED
Sbjct: 344 NLSSLAVLFRLLRQQGLHVLPNVFNKFKDELGNFSEKLIKNVEGMLSLYEATHIMVHGED 523
Query: 121 GLNDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKW 180
L + + L++ + + H A+ + L+ +H + R +S + +
Sbjct: 524 ILEEALAFTTTQLES-ISKQSSHSHALQAKHCLRQTLHKNIPRLEARSYISRYEEDPSHN 700
Query: 181 ICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPC 240
L LAK++ ++++ +++ E KWW++L + ++ +A + W + + +P
Sbjct: 701 ENLLILAKLDFNMLQSLHQKEFGNFSKWWKELDVRSKLPYARDRIAESCFWALGVYYEPQ 880
Query: 241 FSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMK---VSL 297
+S R +TK ++ +IDD +D G +D+L LFT+A+ RW++ +NLP++MK V +
Sbjct: 881 YSTARKVMTKLFVIITVIDDTYDAFGRIDELELFTKALERWDISCLDNLPDYMKFLYVII 1060
Query: 298 SSLYKVTNNFAEMVYKKHGFN-PIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNG 356
LYK E KK G ++ +++ + A++ EA WLN PT EEY+
Sbjct: 1061LDLYKE----IEQEMKKEGREYALNYYVKEFIKYVQAYMTEARWLNDKYQPTLEEYIRIS 1228
Query: 357 IVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQ 416
S G +V + M E + P +I + + RL DD+ + +
Sbjct: 1229TESCGYALVTTTCYIFMGDT---ATEDIFKWVSNGPKMINAAIILCRLMDDIASNEFEQK 1399
Query: 417 NGLDGSYLNC 426
S+L C
Sbjct: 1400RDHVSSFLEC 1429
>TC78150
Length = 612
Score = 147 bits (372), Expect = 6e-36
Identities = 74/123 (60%), Positives = 84/123 (68%), Gaps = 30/123 (24%)
Frame = +1
Query: 280 RWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKIS------------- 326
RWE+ G E LPNFMKV+L++LY +TNNFAEMVYKKHGFNPIDTLK S
Sbjct: 244 RWEITGTEQLPNFMKVALNALYDITNNFAEMVYKKHGFNPIDTLKKSVH*LSVFFI*SYI 423
Query: 327 -----------------WVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHA 369
W+ LLNAF++EAHWLNSG LP E+YLNNGIVSTGVHVVL+HA
Sbjct: 424 *ISFSNG*VYKNICVKQWILLLNAFMEEAHWLNSGHLPRAEDYLNNGIVSTGVHVVLVHA 603
Query: 370 FFL 372
FFL
Sbjct: 604 FFL 612
>BE319697 weakly similar to GP|4508070|gb|A 24349 {Arabidopsis thaliana},
partial (13%)
Length = 613
Score = 119 bits (299), Expect = 2e-27
Identities = 51/68 (75%), Positives = 61/68 (89%)
Frame = +3
Query: 216 KEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFT 275
KE+KF+ Y PLKWY WPMACFTDP FS +R+ELTKPISL+Y+IDDIFDVH TLDQLT+FT
Sbjct: 3 KEVKFSEYQPLKWYTWPMACFTDPNFSEQRIELTKPISLIYVIDDIFDVHATLDQLTIFT 182
Query: 276 EAVNRWEM 283
+AVNR+ +
Sbjct: 183 DAVNRYSI 206
Score = 117 bits (293), Expect = 8e-27
Identities = 51/68 (75%), Positives = 60/68 (88%)
Frame = +2
Query: 280 RWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAH 339
RWE G E LPNFMK++L++LY +TN+FAEMVYKKHGFNPIDTLK SW+ LLNAF++EAH
Sbjct: 407 RWEFTGTEQLPNFMKIALNALYDITNSFAEMVYKKHGFNPIDTLKKSWILLLNAFMEEAH 586
Query: 340 WLNSGILP 347
WLNSG LP
Sbjct: 587 WLNSGHLP 610
>TC84773 weakly similar to GP|17227146|gb|AAL38029.1 linalool synthase
{Perilla frutescens var. crispa}, partial (6%)
Length = 744
Score = 95.9 bits (237), Expect = 3e-20
Identities = 46/123 (37%), Positives = 73/123 (58%)
Frame = +2
Query: 304 TNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVH 363
TN A + K+HG + LK +W+ + AFL+EA W N+G +P + YL+NG++S G
Sbjct: 8 TNEIAYKIQKEHGLTVVSYLKRTWIDIFEAFLEEAKWFNNGCVPNFKTYLDNGVISAGSC 187
Query: 364 VVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSY 423
+ L+HA FL+ G++KET+SI+ + +P + +ILRL DDL + G + S
Sbjct: 188 MALVHATFLI--GDGLSKETMSIMMKSYPRLFTCSGEILRLWDDLGTSTEEQERGDNASS 361
Query: 424 LNC 426
+ C
Sbjct: 362 IQC 370
>TC90477 weakly similar to GP|9864189|gb|AAG01339.1| terpene synthase
{Citrus junos}, partial (12%)
Length = 791
Score = 75.1 bits (183), Expect = 5e-14
Identities = 56/191 (29%), Positives = 89/191 (46%)
Frame = +1
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
++ + LV S DP +ID IQRLGI H+ +EI + L D+ S +L
Sbjct: 238 RRSREALVNSNDPIVTLKMIDSIQRLGIGHHLEDEINIQL--------GRICDWDLSQDL 393
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
+ +L FRLLR G+ +++F + +DV G+++LYEAS L E E+ L
Sbjct: 394 FATSLQFRLLRHNGWTTCSDVFRKFLDKSGNFNESLTKDVWGMLSLYEASYLGTEDEEIL 573
Query: 123 NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWIC 182
S L +P H ++ +L P H ++R ++ K N+
Sbjct: 574 KKAMEFSRARLSELIP-HLSPEVGRNIAKSLTLPKHLRMARLEARNYMEEYSKGSNQIPA 750
Query: 183 LDELAKMNSSI 193
L +LAK++S I
Sbjct: 751 LLKLAKLDSDI 783
>TC80101 weakly similar to GP|9864189|gb|AAG01339.1| terpene synthase
{Citrus junos}, partial (14%)
Length = 679
Score = 73.2 bits (178), Expect = 2e-13
Identities = 41/118 (34%), Positives = 70/118 (58%), Gaps = 3/118 (2%)
Frame = +1
Query: 10 VTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPID---FVSSHELYEVA 66
V +P+ +LID IQRLG+ H+F +EI L + +N ++ + +L+ +A
Sbjct: 307 VNVTNPSREANLIDSIQRLGLYHHFEQEIGELLRH----IDNNHVENGTITLNEDLHSIA 474
Query: 67 LAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND 124
L FRLLRQ GY++ ++F K + + + DV+G+++LYEA+ + I GE+ L+D
Sbjct: 475 LVFRLLRQQGYHILPDVFKKFKNEQGNFKETLVGDVEGMLSLYEATHMRIHGEEILDD 648
>TC82073 weakly similar to GP|14331015|emb|CAC41012. putative chloroplast
terpene synthase {Quercus ilex}, partial (16%)
Length = 654
Score = 70.5 bits (171), Expect = 1e-12
Identities = 41/116 (35%), Positives = 67/116 (57%)
Frame = +2
Query: 4 QVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELY 63
Q K + ++ LID +++LG+ ++F EEI AL++ FL L L+
Sbjct: 308 QEEVKRMIKDENVNILELIDTVKQLGLSYHFEEEIGEALDR-FLSLEKCSGRNNFGRSLH 484
Query: 64 EVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGE 119
E AL FRLLR+ GY ++ ++F+ K + + +D+KG+++LY+AS LS EGE
Sbjct: 485 ETALRFRLLREYGYDISPDIFEKFKDHNGNFKACLVQDIKGMLSLYDASFLSYEGE 652
>BQ149450 weakly similar to GP|5360741|dbj vetispiradiene synthase {Solanum
tuberosum}, partial (22%)
Length = 628
Score = 54.3 bits (129), Expect(2) = 2e-11
Identities = 30/99 (30%), Positives = 57/99 (57%)
Frame = +1
Query: 65 VALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND 124
+A+ FRLLRQ G++V+ +F+ K + + + DV+G+++LYEA+ + + G+D L +
Sbjct: 238 LAVLFRLLRQQGFHVSPNVFNKFKDEQGNFSERLIMDVEGMLSLYEATHVRVHGDDILEE 417
Query: 125 LGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSR 163
+ L++ + H AI V ++L+ +H L R
Sbjct: 418 ALAFTTTHLES-IANQTSHSNAIQVRHSLRQALHKNLPR 531
Score = 32.3 bits (72), Expect(2) = 2e-11
Identities = 17/38 (44%), Positives = 22/38 (57%)
Frame = +2
Query: 12 SEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLIL 49
+E P HLID I RLGI ++F EI+ L F I+
Sbjct: 74 TEMPLAKIHLIDSICRLGISYHFEHEIEEVLLHIFTII 187
>TC85843 weakly similar to GP|3687297|emb|CAA06614.1 5-epi-aristolochene
synthase {Capsicum annuum}, partial (14%)
Length = 563
Score = 55.8 bits (133), Expect = 3e-08
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 1/114 (0%)
Frame = +1
Query: 4 QVHKKLVTSED-PTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
+V LV D P L+D I RLG+ ++F +EI L++ N + + L
Sbjct: 205 EVRNMLVAKTDEPFAKVKLVDSICRLGVSYHFEKEIDDVLQQVHKSCVENG-EVILEDNL 381
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSI 116
+A+ FR+LRQ G+YV+ +F K + + DV+G+++LYEA + +
Sbjct: 382 CALAVLFRVLRQQGFYVSPNVFTKFKDKQGNFNETLITDVEGMLSLYEAXHMIV 543
>BI270658 weakly similar to PIR|T09672|T096 ent-kaurene synthase B (EC
2.5.1.-) - winter squash, partial (10%)
Length = 261
Score = 43.1 bits (100), Expect = 2e-04
Identities = 25/71 (35%), Positives = 38/71 (53%)
Frame = +3
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLR 73
D H+ID ++RLGI H+F EEI+ L++ + D + A+AFR+LR
Sbjct: 18 DVYARLHMIDSLERLGINHHFKEEIQSVLDETYRYWLQRAEDIFL--DPTTCAMAFRMLR 191
Query: 74 QGGYYVNAELF 84
GY V++ F
Sbjct: 192LNGYDVSSXPF 224
>AL373351 weakly similar to PIR|T52059|T520 ent-kaurene synthase (EC 2.5.1.-)
[imported] - Arabidopsis thaliana, partial (7%)
Length = 356
Score = 40.8 bits (94), Expect = 0.001
Identities = 19/76 (25%), Positives = 36/76 (47%)
Frame = +1
Query: 298 SSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGI 357
S++ F E K+ G N + L W ++ + EA W S +PT + Y+ N
Sbjct: 7 SAMRSTICEFGEKSVKRQGRNVTENLIKIWSNMIQSMFIEAEWSRSKTIPTIDVYMENAC 186
Query: 358 VSTGVHVVLIHAFFLM 373
+ G+ +++ A +L+
Sbjct: 187 ETFGLEPLILTALYLL 234
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,064,974
Number of Sequences: 36976
Number of extensions: 202481
Number of successful extensions: 968
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 941
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 949
length of query: 426
length of database: 9,014,727
effective HSP length: 99
effective length of query: 327
effective length of database: 5,354,103
effective search space: 1750791681
effective search space used: 1750791681
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142394.1


