

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142224.5 - phase: 0 /pseudo
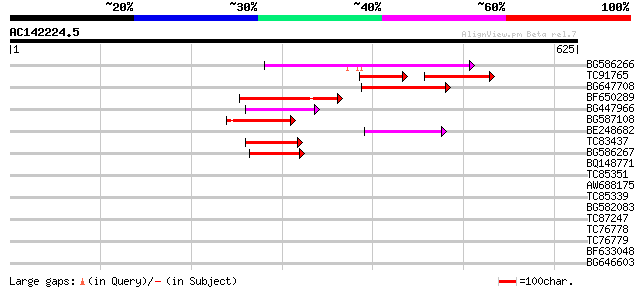
(625 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 154 7e-38
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 71 3e-23
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 100 2e-21
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 74 2e-13
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 65 8e-11
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 65 1e-10
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 64 1e-10
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 64 2e-10
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 53 4e-07
BQ148771 28 0.28
TC85351 32 0.56
AW688175 32 0.56
TC85339 32 0.56
BG582083 32 0.95
TC87247 similar to GP|9714501|emb|CAC01441.1 putative fatty acid... 30 2.8
TC76778 homologue to GP|18158221|gb|AAL62332.1 aminotransferase ... 30 3.6
TC76779 similar to GP|18032028|gb|AAL47679.1 aminotransferase 1 ... 30 3.6
BF633048 similar to GP|11994760|d contains similarity to pheroph... 30 3.6
BG646603 homologue to GP|20197786|gb| expressed protein {Arabido... 29 6.2
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 154 bits (390), Expect = 7e-38
Identities = 90/246 (36%), Positives = 137/246 (55%), Gaps = 14/246 (5%)
Frame = -3
Query: 281 VSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTKG 340
++ CN YKI++K+L+ R++ +L IS SQ+AFVP R I +N L ++LHY++
Sbjct: 760 IAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLRQSGAK 581
Query: 341 KEGNIALKQDVSKAFDRIKWSYLQAVMEKK-----WDSQMFGLTGSC------NV*P--- 386
K ++A+K D++KA+DRI W++L+ V+ + W S + + N P
Sbjct: 580 KHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLINGGPQGR 401
Query: 387 IDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSF 446
+ P G+RQG P SPYL+I+C+E L+ + G + G ++ R P I LLFADD+
Sbjct: 400 VLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLLFADDTM 221
Query: 447 LFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMW 506
F K+ S IL +I+ Y AASG+ IN KS+I FS T + L + +
Sbjct: 220 FFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELKIAKEGG 41
Query: 507 HGKYLG 512
GKYLG
Sbjct: 40 TGKYLG 23
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 70.9 bits (172), Expect(2) = 3e-23
Identities = 39/77 (50%), Positives = 51/77 (65%)
Frame = +3
Query: 458 ILKNILYTYEAASGQAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMV 517
I+KNIL YE SG+AI+ RKS I SRN I +IT++LGV + KYLGLPSM+
Sbjct: 336 IMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYILGVQFMLGTCKYLGLPSMI 515
Query: 518 ERDKKSIFSFIKERIWK 534
RD+ + FS IK +W+
Sbjct: 516 GRDRTTTFSSIKGGVWQ 566
Score = 56.2 bits (134), Expect(2) = 3e-23
Identities = 26/53 (49%), Positives = 35/53 (65%)
Frame = +2
Query: 386 PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQ 438
PI P G++QG SPY++IIC EGL+ I H + G HGT I RG+P ++Q
Sbjct: 155 PIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTSI*RGAPPVSQ 313
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 100 bits (249), Expect = 2e-21
Identities = 43/98 (43%), Positives = 67/98 (67%)
Frame = +1
Query: 389 PHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLF 448
P G+RQG P SPYL+I+C+ L+ +K N +HG ++ R P IT LLFADDS LF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 449 YKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRN 486
+A ++E + +L++Y++ASGQ +N+ KS +++S+N
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQN 297
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 73.9 bits (180), Expect = 2e-13
Identities = 38/115 (33%), Positives = 70/115 (60%), Gaps = 1/115 (0%)
Frame = +3
Query: 254 FPPNLID-TNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQA 312
F P +I+ T + L+ K S+K+ P++ C+VIYKI+SK+L +R++ +L+ +S++Q+
Sbjct: 177 FMPKIINCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQS 356
Query: 313 AFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVM 367
AFV R I +N + E++ KG +K D+ KA+D +W +++ +M
Sbjct: 357 AFVKGRVIFDNIILSHELV--KSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLM 515
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 65.1 bits (157), Expect = 8e-11
Identities = 37/81 (45%), Positives = 49/81 (59%)
Frame = +2
Query: 261 TNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFI 320
T I LI K P + K P+SLCNV+ KI++KV+ANR+KQ L I Q+AFV R I
Sbjct: 434 TFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIANRVKQTLPDVIDVEQSAFVQGRLI 613
Query: 321 LENALTYFEVLHYMKCKTKGK 341
+NAL + V + + KGK
Sbjct: 614 TDNALIAWSVSIG*RXRRKGK 676
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 64.7 bits (156), Expect = 1e-10
Identities = 37/76 (48%), Positives = 47/76 (61%)
Frame = +3
Query: 240 LIKLL*KPIVSEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRL 299
L+K++ ++ G L TNI LI K RP M L P+SLCNV YKI+SKVL RL
Sbjct: 447 LLKMV-NSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGYKIISKVLCQRL 623
Query: 300 KQILHKCISDSQAAFV 315
K L IS++Q+AFV
Sbjct: 624 KVCLPSLISETQSAFV 671
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 64.3 bits (155), Expect = 1e-10
Identities = 30/90 (33%), Positives = 52/90 (57%)
Frame = +3
Query: 392 GIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKA 451
G++QG P +P+L+++ +EG++ +K+ N L G + RG ++ L +ADD+
Sbjct: 27 GLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGMP 206
Query: 452 YVSEDTILKNILYTYEAASGQAINYRKSSI 481
V LK +L +E ASG +N+ KSS+
Sbjct: 207 TVDNLWTLKALLQGFEMASGLKVNFHKSSL 296
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 63.9 bits (154), Expect = 2e-10
Identities = 32/62 (51%), Positives = 44/62 (70%)
Frame = +2
Query: 261 TNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFI 320
T IALI K+D P+ + P+SL +YKIL K+LANRL+ ++ ISD+Q+AFV +R I
Sbjct: 59 TFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKNRQI 238
Query: 321 LE 322
LE
Sbjct: 239 LE 244
Score = 39.7 bits (91), Expect = 0.004
Identities = 34/132 (25%), Positives = 60/132 (44%), Gaps = 3/132 (2%)
Frame = +2
Query: 436 ITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIA---FSRNTDSIGH 492
++ L FA+D+ L + L+ L + A SG +N+ KS + + + S
Sbjct: 542 VSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFHKSGLVCVNIAPSWLSEAA 721
Query: 493 HNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSRAGKEVL 552
++ +G V + YLG+P + S + I RI + W++ LS G+ VL
Sbjct: 722 SVLSWKVGKVPFL----YLGMPIEGNSRRLSFWEPIVNRIKARLTGWNSRFLSFGGRLVL 889
Query: 553 LKSIAQYIPTYS 564
LKS+ + Y+
Sbjct: 890 LKSVLTSLSVYA 925
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 52.8 bits (125), Expect = 4e-07
Identities = 28/61 (45%), Positives = 39/61 (63%)
Frame = +1
Query: 265 LIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFILENA 324
L+ K K + P+SLCNV YKI+SKVL+ RLK +L I+++QAAF + I +N
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 325 L 325
L
Sbjct: 781 L 783
>BQ148771
Length = 680
Score = 28.1 bits (61), Expect(2) = 0.28
Identities = 16/43 (37%), Positives = 24/43 (55%)
Frame = -3
Query: 521 KKSIFSFIKERIWKNIQNWSAWSLSRAGKEVLLKSIAQYIPTY 563
KKS FS + ++ + NW A LS A + L KS+ + +P Y
Sbjct: 615 KKSRFS-VYYQVHVMLANWKANHLSLARRVTLAKSVIEAVPLY 490
Score = 23.9 bits (50), Expect(2) = 0.28
Identities = 10/21 (47%), Positives = 13/21 (61%)
Frame = -2
Query: 508 GKYLGLPSMVERDKKSIFSFI 528
GKY G+P E K+ IFS +
Sbjct: 652 GKYXGVPC*EEHQKEPIFSIL 590
>TC85351
Length = 720
Score = 32.3 bits (72), Expect = 0.56
Identities = 18/53 (33%), Positives = 31/53 (57%), Gaps = 3/53 (5%)
Frame = +1
Query: 95 VVLHLL---CLPFLQLLMWKTVHDLWLSCGVFGALVSHAYGNKNLLMLKHLVF 144
+V HL+ C+PFL++LM +W+S G+ ++S Y NL+ L ++F
Sbjct: 184 IVPHLI*HHCIPFLEMLMLALQGQMWVSIGI---VLSGNYLVNNLVFLTLVIF 333
>AW688175
Length = 656
Score = 32.3 bits (72), Expect = 0.56
Identities = 18/53 (33%), Positives = 31/53 (57%), Gaps = 3/53 (5%)
Frame = +2
Query: 95 VVLHLL---CLPFLQLLMWKTVHDLWLSCGVFGALVSHAYGNKNLLMLKHLVF 144
+V HL+ C+PFL++LM +W+S G+ ++S Y NL+ L ++F
Sbjct: 452 IVPHLI*HHCIPFLEMLMLALQGQMWVSIGI---VLSGNYLVNNLVFLTLVIF 601
>TC85339
Length = 786
Score = 32.3 bits (72), Expect = 0.56
Identities = 18/53 (33%), Positives = 31/53 (57%), Gaps = 3/53 (5%)
Frame = +1
Query: 95 VVLHLL---CLPFLQLLMWKTVHDLWLSCGVFGALVSHAYGNKNLLMLKHLVF 144
+V HL+ C+PFL++LM +W+S G+ ++S Y NL+ L ++F
Sbjct: 397 IVPHLI*HHCIPFLEMLMLALQGQMWVSIGI---VLSGNYLVNNLVFLTLVIF 546
>BG582083
Length = 665
Score = 31.6 bits (70), Expect = 0.95
Identities = 13/18 (72%), Positives = 16/18 (88%)
Frame = +3
Query: 292 SKVLANRLKQILHKCISD 309
S +L NRLKQ+LHKCIS+
Sbjct: 210 SFLLVNRLKQVLHKCISE 263
>TC87247 similar to GP|9714501|emb|CAC01441.1 putative fatty acid elongase
{Zea mays}, partial (91%)
Length = 2102
Score = 30.0 bits (66), Expect = 2.8
Identities = 16/54 (29%), Positives = 26/54 (47%), Gaps = 1/54 (1%)
Frame = +3
Query: 484 SRNTDSIGHHNITHLLGVVESMW-HGKYLGLPSMVERDKKSIFSFIKERIWKNI 536
S T + N+ ++E +W K L PS +E KS++ KER W+ +
Sbjct: 555 SELTGTFSDENLAFQKKILEKVWFRSKDLFAPSNIECSTKSLYG*SKERSWRKL 716
>TC76778 homologue to GP|18158221|gb|AAL62332.1 aminotransferase 2 {Cucumis
melo}, partial (28%)
Length = 1210
Score = 29.6 bits (65), Expect = 3.6
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = +3
Query: 529 KERIWKNIQNWSAWSLSR 546
K+ WK QNW++W L R
Sbjct: 159 KQSCWKGFQNWTSWQLER 212
>TC76779 similar to GP|18032028|gb|AAL47679.1 aminotransferase 1 {Cucumis
melo}, complete
Length = 1491
Score = 29.6 bits (65), Expect = 3.6
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = +2
Query: 529 KERIWKNIQNWSAWSLSR 546
K+ WK QNW++W L R
Sbjct: 1142 KQSCWKGFQNWTSWQLER 1195
>BF633048 similar to GP|11994760|d contains similarity to
pherophorin~gene_id:T5M7.14 {Arabidopsis thaliana},
partial (15%)
Length = 510
Score = 29.6 bits (65), Expect = 3.6
Identities = 11/22 (50%), Positives = 16/22 (72%)
Frame = -1
Query: 65 CICLCTGSLLLIAGER*IFAES 86
CICLC+ +L +A R +FAE+
Sbjct: 117 CICLCSSFILFLANSRSLFAEA 52
>BG646603 homologue to GP|20197786|gb| expressed protein {Arabidopsis
thaliana}, partial (20%)
Length = 672
Score = 28.9 bits (63), Expect = 6.2
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 3/46 (6%)
Frame = +3
Query: 102 LPFLQLLMWKTVHDLWLSCGVFG---ALVSHAYGNKNLLMLKHLVF 144
LP LQ L + +LW SCG+ G A+ + +N L+LKH +F
Sbjct: 462 LPQLQFLWPRLAGELWPSCGLPGIPSAIKVLSSIFQNALLLKHCLF 599
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.342 0.149 0.502
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,781,569
Number of Sequences: 36976
Number of extensions: 406791
Number of successful extensions: 3217
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 2375
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 795
Number of HSP's gapped (non-prelim): 2539
length of query: 625
length of database: 9,014,727
effective HSP length: 102
effective length of query: 523
effective length of database: 5,243,175
effective search space: 2742180525
effective search space used: 2742180525
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC142224.5


