

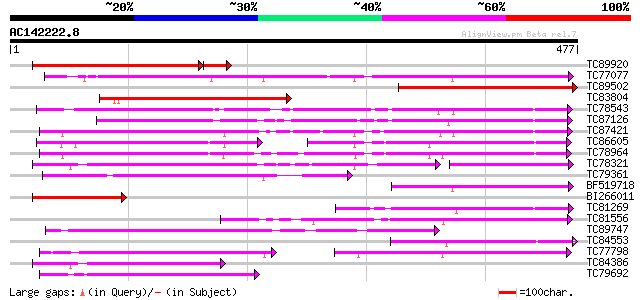
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.8 - phase: 0
(477 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89920 similar to GP|12039393|gb|AAG46179.1 putative sugar tran... 275 3e-83
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 218 3e-57
TC89502 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-l... 208 4e-54
TC83804 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-l... 191 7e-49
TC78543 similar to PIR|T14545|T14545 probable sugar transporter ... 185 4e-47
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 174 5e-44
TC87421 sugar transporter 168 4e-42
TC86605 similar to PIR|T01853|T01853 probable hexose transport p... 100 8e-41
TC78964 weakly similar to PIR|T00450|T00450 probable monosacchar... 153 1e-37
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 151 5e-37
TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 122 3e-28
BF519718 weakly similar to PIR|G84564|G84 probable sugar transpo... 115 5e-26
BI266011 similar to GP|9759246|dbj| sugar transporter-like prote... 114 1e-25
TC81269 similar to GP|23504385|emb|CAC00697. putative sugar tran... 113 1e-25
TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar tran... 113 1e-25
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 106 2e-23
TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90 ... 102 3e-22
TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 100 1e-21
TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 99 3e-21
TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 96 2e-20
>TC89920 similar to GP|12039393|gb|AAG46179.1 putative sugar transporter
protein {Oryza sativa}, partial (33%)
Length = 654
Score = 275 bits (704), Expect(2) = 3e-83
Identities = 143/144 (99%), Positives = 143/144 (99%)
Frame = +2
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L FLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS
Sbjct: 152 LPFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 331
Query: 80 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 139
VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY
Sbjct: 332 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 511
Query: 140 IAETAPTPIRGQLVSLKEFFIVIG 163
IAETAPTPIRGQLVSLKEFFIVIG
Sbjct: 512 IAETAPTPIRGQLVSLKEFFIVIG 583
Score = 51.6 bits (122), Expect(2) = 3e-83
Identities = 23/23 (100%), Positives = 23/23 (100%)
Frame = +3
Query: 164 IVAGYGLGSLLVDTVAGWRYMFG 186
IVAGYGLGSLLVDTVAGWRYMFG
Sbjct: 585 IVAGYGLGSLLVDTVAGWRYMFG 653
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 218 bits (556), Expect = 3e-57
Identities = 154/467 (32%), Positives = 235/467 (49%), Gaps = 22/467 (4%)
Frame = +1
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDA----VEIGLLTSGSLYGALIGSVLAFNI 85
+L GYDIG S A I I+ DL +E+ LL ++Y +LIGS LA
Sbjct: 145 ILLGYDIGVMSGAAIYIKR---------DLKVSDGKIEV-LLGIINIY-SLIGSCLAGRT 291
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
+D++GRR ++ A ++ VGAL+ F+PN+ L+ GR V G+GIG A+ AP+Y AE +P
Sbjct: 292 SDWIGRRYTIVFAGAIFFVGALLMGFSPNYNFLMFGRFVAGVGIGYALMIAPVYTAEVSP 471
Query: 146 TPIRGQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLP 203
RG L S E FI GI+ GY + GWR M G+ + +VI+ G+ +P
Sbjct: 472 ASSRGFLTSFPEVFINSGILLGYISNYAFSKLSLKLGWRMMLGVGAIPSVILAVGVLAMP 651
Query: 204 ASPRWILLR-----AIQKKGDLQTLKDTAIRSLCQL-QGRTFHDSAPQQVDEIMAEFSYL 257
SPRW+++R AI+ K+ A L ++ Q + V E+ + +
Sbjct: 652 ESPRWLVMRGRLGDAIKVLNKTSDSKEEAQLRLAEIKQAAGIPEDCNDDVVEVKVKNTGE 831
Query: 258 GEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLA 314
G ++ L R ++ + G+ FQQ + V + PT G +
Sbjct: 832 GVWKELFLYP--TPAVRHIVIAALGIHFFQQASGVDAVVLYSPT------IFKKAGINGD 987
Query: 315 ADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN--- 371
+I +G K + VA ++DR GRRPLLL V G+V+SL L +D+
Sbjct: 988 THLLIATIAVGFVKTLFILVATFMLDRYGRRPLLLTSVGGMVVSLLTLAVSLTIIDHSNT 1167
Query: 372 ----AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVT 427
A L++ +L YV + I GP+ W+ +EIFPLRLR +G + V+VN + +++
Sbjct: 1168KLNWAIGLSIATVLSYVATFSIGAGPITWVYSSEIFPLRLRAQGAACGVVVNRVTSGVIS 1347
Query: 428 FAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
F L + G F++F IA +F Y ++PET+G TLEE+EA
Sbjct: 1348MTFLSLSKGITIGGAFFLFGGIATIGWIFFYIMLPETQGKTLEEMEA 1488
>TC89502 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 595
Score = 208 bits (529), Expect = 4e-54
Identities = 105/150 (70%), Positives = 125/150 (83%)
Frame = +2
Query: 328 KLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVVGLLLYVGCY 387
KL+MT VAV+ VD LGRRPLL+GGVSGI +SL LL +YY FL ++AV LLLYVGCY
Sbjct: 2 KLVMTSVAVLKVDDLGRRPLLIGGVSGIALSLVLLSAYYKFLGGLPIVAVAALLLYVGCY 181
Query: 388 QISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFS 447
QISFGP+ WLM++EIFPLR RG+G+S+AVL NFAANA+VTFAFSPLK+ LGA LF +F+
Sbjct: 182 QISFGPISWLMVSEIFPLRTRGRGISMAVLTNFAANAVVTFAFSPLKEYLGAENLFLLFA 361
Query: 448 AIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
AIA+ SLVFI VPETKGL+LEEIE+K L
Sbjct: 362 AIALVSLVFIVTSVPETKGLSLEEIESKIL 451
Score = 28.9 bits (63), Expect = 4.5
Identities = 28/108 (25%), Positives = 46/108 (41%), Gaps = 8/108 (7%)
Frame = +2
Query: 67 LTSGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAP--------NFPLL 118
L G + G + VL FLG + VAAL+ VG +F P FPL
Sbjct: 59 LLIGGVSGIALSLVLLSAYYKFLGGLPIVAVAALLLYVGCYQISFGPISWLMVSEIFPLR 238
Query: 119 VIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVA 166
GR GI + + + A + A +P++ + + + F++ +A
Sbjct: 239 TRGR---GISMAVLTNFAANAVVTFAFSPLK-EYLGAENLFLLFAAIA 370
>TC83804 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-like protein
{Arabidopsis thaliana}, partial (29%)
Length = 721
Score = 191 bits (484), Expect = 7e-49
Identities = 95/171 (55%), Positives = 129/171 (74%), Gaps = 9/171 (5%)
Frame = +3
Query: 76 LIGSVLAFNIA---DFLG------RRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFG 126
L+G++L+ + +FLG R+R+L+ AAL+Y++G+ ITA AP +L+ GRL++G
Sbjct: 171 LLGTILSLLLPTKRNFLGLLLFFRRKRQLIGAALLYVLGSAITATAPELGVLLAGRLLYG 350
Query: 127 IGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFG 186
+GIGLAMH AP+YIAET P+ IRG LVSLKE FIV+GI+ GY +GS + TV GWR+M+G
Sbjct: 351 LGIGLAMHGAPLYIAETCPSQIRGTLVSLKELFIVLGILLGYFVGSFQISTVGGWRFMYG 530
Query: 187 ISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGR 237
S+P+AV+MG GMW LP SPRW+LL A+Q KG Q LK+ AI SL +L+GR
Sbjct: 531 FSAPLAVLMGLGMWTLPESPRWLLLNAVQGKGSFQDLKEKAIVSLGKLRGR 683
>TC78543 similar to PIR|T14545|T14545 probable sugar transporter protein -
beet, partial (93%)
Length = 1653
Score = 185 bits (469), Expect = 4e-47
Identities = 133/462 (28%), Positives = 237/462 (50%), Gaps = 11/462 (2%)
Frame = +1
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLA 82
L A G + FG+ G TS +I IT L E L S S GA++G++ +
Sbjct: 163 LIVALGPIQFGFTAGYTSPTQSAI-------ITDLGLSVSEFSLFGSLSNVGAMVGAIAS 321
Query: 83 FNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAE 142
IA+++GR+ L++A++ ++G L+ +FA + L +GRL+ G G+G+ + P+YIAE
Sbjct: 322 GQIAEYMGRKGSLMIASIPNIIGWLMISFANDSSFLYMGRLLEGFGVGIISYTVPVYIAE 501
Query: 143 TAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWL 202
+P +RG LVS+ + + +GI+ Y LG L V+ WR++ + ++ G++++
Sbjct: 502 ISPQNLRGSLVSVNQLSVTLGIMLAYLLG-LFVE----WRFLAILGIIPCTLLIPGLFFI 666
Query: 203 PASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEND 262
P SPRW+ + ++ + SL L+G F +V+EI +
Sbjct: 667 PESPRWLAKMGMTEEFE---------NSLQVLRG--FETDISVEVNEIKTAVASANRRTT 813
Query: 263 VTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSI 322
V E+ + + L+I GL++ QQ+ + ++ + G S + AT
Sbjct: 814 VRFSELKQRRYWLPLMIGIGLLVLQQLSGINGVLFYSSTIFQ---NAGISSSDVAT---F 975
Query: 323 LLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISL------FLLGSYYIFLDNA---- 372
+G +++ T + + + D+ GRR LL+ S + +SL F L YYI D++
Sbjct: 976 GVGAVQVLATTLTLWLADKSGRRLLLIVSSSAMTLSLLVVSISFYLKDYYISADSSLYGI 1155
Query: 373 -AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFS 431
++L+V G+++ V + + G M W++++EI P+ ++G S A L N+ + L+T +
Sbjct: 1156LSLLSVAGVVVMVIAFSLGMGAMPWIIMSEILPINIKGLAGSFATLANWFFSWLITLTAN 1335
Query: 432 PLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
L D G F I++ + ++ F+ VPETKG TLEEI+
Sbjct: 1336LLLDWSSGG-TFTIYTVVCAFTVGFVAIWVPETKGKTLEEIQ 1458
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 174 bits (442), Expect = 5e-44
Identities = 122/410 (29%), Positives = 217/410 (52%), Gaps = 10/410 (2%)
Frame = +3
Query: 74 GALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAM 133
GA++G++ + IA+++GR+ L++A++ ++G L +FA + L +GRL+ G G+G+
Sbjct: 6 GAMVGAIASGQIAEYVGRKGSLMIASIPNIIGWLAISFAKDSSFLFMGRLLEGFGVGIIS 185
Query: 134 HAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAV 193
+ P+YIAE AP +RG L S+ + + IGI+ Y LG A WR + +
Sbjct: 186 YVVPVYIAEIAPENMRGSLGSVNQLSVTIGIMLAYLLG-----LFANWRVLAILGILPCT 350
Query: 194 IMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAE 253
++ G++++P SPRW + K G ++ +T+++ L R F +V EI
Sbjct: 351 VLIPGLFFIPESPRW-----LAKMGMMEEF-ETSLQVL-----RGFDTDISVEVHEIKKA 497
Query: 254 FSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSL 313
+ G+ + ++ R + L + GL++ QQ+ + +G S N G S
Sbjct: 498 VASNGKRATIRFADLQRKRYWFPLSVGIGLLVLQQLSGI--NGVLFYSTSIFAN-AGISS 668
Query: 314 AADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLG-SYYI----- 367
+ AT + LG ++I TGVA +VD+ GRR LL+ S + SL ++ ++Y+
Sbjct: 669 SNAAT---VGLGAIQVIATGVATWLVDKSGRRVLLIISSSLMTASLLVVSIAFYLEGVVE 839
Query: 368 ----FLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAAN 423
+ +++VVGL++ V + + GP+ WL+++EI P+ ++G S A + N+
Sbjct: 840 KDSQYFSILGIISVVGLVVMVIGFSLGLGPIPWLIMSEILPVNIKGLAGSTATMANWLVA 1019
Query: 424 ALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
++T + L G F I++ +A ++VF VPETKG TLEEI+
Sbjct: 1020WIITMTANLLLTWSSGG-TFLIYTVVAAFTVVFTSLWVPETKGRTLEEIQ 1166
>TC87421 sugar transporter
Length = 1753
Score = 168 bits (426), Expect = 4e-42
Identities = 138/478 (28%), Positives = 229/478 (47%), Gaps = 30/478 (6%)
Frame = +2
Query: 26 AFGGLLFGYDIGATSSAT-------------ISIQSSSLSGITWYDLDAVEIGLLTSGSL 72
A GGL+FGYDIG + T ++ S + D+ + + TS
Sbjct: 119 AMGGLIFGYDIGISGGVTSMDPFLKKFFPAVYRKKNKDKSTNQYCQYDSQTLTMFTSSLY 298
Query: 73 YGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLA 132
AL+ S++A I GR+ +L L++LVGALI FA + +L++GR++ G GIG A
Sbjct: 299 LAALLSSLVASTITRRFGRKLSMLFGGLLFLVGALINGFANHVWMLIVGRILLGFGIGFA 478
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVA--GWRYMFGISSP 190
P+Y++E AP RG L + I IGI+ L GWR G +
Sbjct: 479 NQPVPLYLSEMAPYKYRGALNIGFQLSITIGILVANVLNYFFAKIKGGWGWRLSLGGAMV 658
Query: 191 VAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEI 250
A+I+ G LP +P ++ ++GD +D A L +++G D + D +
Sbjct: 659 PALIITIGSLVLPDTPNSMI-----ERGD----RDGAKAQLKRIRG--IEDVDEEFNDLV 805
Query: 251 MAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCIN 307
A + + EN + + K R L ++ + FQQ I + + P N
Sbjct: 806 AASEASMQVEN--PWRNLLQRKYRPQLTMAVLIPFFQQFTGINVIMFYAPV------LFN 961
Query: 308 PTGFSLAADATRVS-ILLGVFKLIMTGVAVVVVDRLGRRPLLL-GGVSGIVISLFL---L 362
GF DA+ +S ++ GV ++ T V++ VD+ GRR L L GG ++ + + +
Sbjct: 962 SIGFK--DDASLMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGAQMLICQVAVAAAI 1135
Query: 363 GSYYIFLDNA-------AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIA 415
G+ + N A++ V+ + +YV + S+GP+GWL+ +EIFPL +R S+
Sbjct: 1136GAKFGTSGNPGNLPEWYAIVVVLFICIYVAGFAWSWGPLGWLVPSEIFPLEIRSAAQSVN 1315
Query: 416 VLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
V VN LV F + + G LF F+ + ++++F++PETKG+ +EE++
Sbjct: 1316VSVNMLFTFLVAQVFLIMLCHMKFG-LFLFFAFFVLVMSIYVFFLLPETKGIPIEEMD 1486
>TC86605 similar to PIR|T01853|T01853 probable hexose transport protein
F9D12.17 - Arabidopsis thaliana, partial (91%)
Length = 3111
Score = 99.8 bits (247), Expect(2) = 8e-41
Identities = 74/234 (31%), Positives = 113/234 (47%), Gaps = 12/234 (5%)
Frame = +3
Query: 251 MAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCIN 307
+ E S + +E + + R LVIS L++FQQ I + + P N
Sbjct: 2061 LVEASRVAKEVKHPFRNLLKRNNRPQLVISIALMIFQQFTGINAIMFYAPV------LFN 2222
Query: 308 PTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGG---------VSGIVIS 358
GF A A +++ G +I T V++ VD+LGRR LLL V IV+
Sbjct: 2223 TLGFKNDA-ALYSAVITGAINVISTIVSIYSVDKLGRRKLLLEAGVQMLLSQMVIAIVLG 2399
Query: 359 LFLLGSYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLV 418
+ + A L VV + ++V + S+GP+ WL+ +EIFPL R G S+ V V
Sbjct: 2400 IKVKDHSEELSKGYAALVVVMVCIFVSAFAWSWGPLAWLIPSEIFPLETRSAGQSVTVCV 2579
Query: 419 NFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
NF A++ AF + GI F+ FS + F++F+VPETK + +EE+
Sbjct: 2580 NFLFTAVIAQAFLSMLCYFKFGI-FFFFSGWILFMSTFVFFLVPETKNVPIEEM 2738
Score = 85.9 bits (211), Expect(2) = 8e-41
Identities = 58/207 (28%), Positives = 98/207 (47%), Gaps = 17/207 (8%)
Frame = +2
Query: 23 LFPAFGGLLFGYDIGATSSA------------TISIQSSSLSGI--TWYDLDAVEIGLLT 68
+ A GGL+FGYD+G + T+ Q++ G + D + L T
Sbjct: 1367 IMAATGGLMFGYDVGVSGGVASMPPFLKKFFPTVLRQTTESDGSESNYCKYDNQGLQLFT 1546
Query: 69 SGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIG 128
S L + A LGRR +L+A ++ G + A A N +L++GR++ G G
Sbjct: 1547 SSLYLAGLTVTFFASYTTRVLGRRLTMLIAGFFFIAGVSLNASAQNLLMLIVGRVLLGCG 1726
Query: 129 IGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVA---GWRYMF 185
IG A A P++++E AP+ IRG L L + I +GI+ L + + + GWR
Sbjct: 1727 IGFANQAVPVFLSEIAPSRIRGALNILFQLDITLGILYA-NLVNYATNKIKGHWGWRISL 1903
Query: 186 GISSPVAVIMGFGMWWLPASPRWILLR 212
G+ A+++ G + + +P ++ R
Sbjct: 1904 GLGGIPALLLTLGAYLVVDTPNSLIER 1984
>TC78964 weakly similar to PIR|T00450|T00450 probable monosaccharide
transport protein T14N5.7 - Arabidopsis thaliana,
partial (98%)
Length = 1771
Score = 153 bits (387), Expect = 1e-37
Identities = 136/474 (28%), Positives = 218/474 (45%), Gaps = 27/474 (5%)
Frame = +1
Query: 26 AFGGLLFGYDIGATSSAT-------------ISIQSSSLSGITWYDLDAVEIGLLTSGSL 72
A GG LFGYD+G + T + + L + D + L TS
Sbjct: 130 ALGGSLFGYDLGVSGGVTSMDDFLEKFFPDVYRKKHAHLKETDYCKYDNQVLTLFTSSLY 309
Query: 73 YGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLA 132
+ AL+ + A + GR+ ++V AL +L+GA++ A A N P L+IGR+ G GIG
Sbjct: 310 FSALVMTFFASYLTRNKGRKATIIVGALSFLIGAILNAAAQNIPTLIIGRVFLGGGIGFG 489
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTV--AGWRYMFGISSP 190
A P+Y++E AP RG + L +F GI+ L + D + GWR G++
Sbjct: 490 NQAVPLYLSEMAPASSRGAVNQLFQFTTCAGILIA-NLVNYFTDKIHPHGWRISLGLAGI 666
Query: 191 VAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEI 250
AV+M G + +P ++ ++G L D A + L +++G D+ E
Sbjct: 667 PAVLMLLGGIFCAETP-----NSLVEQGRL----DEARKVLEKVRGTKNVDAE----FED 807
Query: 251 MAEFSYLGEENDVTLGEMFRGKCRKALVISA-GLVLFQQI---QTVTDHGPTKCSLLRCI 306
+ + S L + + + K R L+I A G FQQ+ ++ + P
Sbjct: 808 LKDASELAQAVKSPFKVLLKRKYRPQLIIGALGTPAFQQLTGNNSILFYAPV------IF 969
Query: 307 NPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGG----VSGIVISLFLL 362
+GF A A S + L+ T +++ +VD+ G R L + ++I+ +L
Sbjct: 970 QSSGFGSNA-ALFSSFITNGALLVATVISMFLVDKFGTRKFFLEAGFEMICCMIITAVVL 1146
Query: 363 ----GSYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLV 418
G + V+ + +V Y S+GP+GWL+ +E+FPL +R SIAV V
Sbjct: 1147AVEFGHGKELSKGISAFLVIMIFWFVLAYGRSWGPLGWLVPSELFPLEIRSAAQSIAVCV 1326
Query: 419 NFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
N ALV F L GI F +F + V VF++F++PETK + +EEI
Sbjct: 1327NMIFTALVAQLFLLSLCHLKYGI-FLLFGGLIVVMSVFVFFLLPETKQVPIEEI 1485
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 151 bits (382), Expect = 5e-37
Identities = 109/350 (31%), Positives = 180/350 (51%), Gaps = 7/350 (2%)
Frame = +2
Query: 20 LRFLFPA-FGGLLFGYDIGATSSATISIQSS--SLSGITWYDLDAVEIGLLTSGSLYGAL 76
LR F A GGLLFGYD G S A + I+ ++ TW + S ++ GA+
Sbjct: 224 LRLAFSAGIGGLLFGYDTGVISGALLYIRDEFPAVEKKTWLQ------EAIVSTAIAGAI 385
Query: 77 IGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAA 136
IG+ + I D GR+ ++VA ++L+G++I A APN L++GR+ G+G+G+A A+
Sbjct: 386 IGAAIGGWINDRFGRKVSIIVADTLFLLGSIILAAAPNPATLIVGRVFVGLGVGMASMAS 565
Query: 137 PMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMG 196
P+YI+E +PT +RG LVSL F I G Y + WR+M G+++ AVI
Sbjct: 566 PLYISEASPTRVRGALVSLNSFLITGGQFLSYLINLAFTKAPGTWRWMLGVAAAPAVIQI 745
Query: 197 FGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSY 256
M LP SPRW + +KG + K ++ + +++ +D+ Q + E +
Sbjct: 746 VLMLSLPESPRW-----LYRKGKEEEAK-VILKKIYEVED---YDNEIQALKESVE--ME 892
Query: 257 LGEENDVTLGEMFR-GKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFS 312
L E +++ ++ + R+ L GL FQQ I TV + P+ L GF+
Sbjct: 893 LKETEKISIMQLVKTTSVRRGLYAGVGLAFFQQFTGINTVMYYSPSIVQL------AGFA 1054
Query: 313 LAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLL 362
A +S++ + +++ +D+ GR+ L L ++G+V+SL LL
Sbjct: 1055SKRTALLLSLITSGLNAFGSILSIYFIDKTGRKKLALISLTGVVLSLTLL 1204
Score = 70.9 bits (172), Expect = 1e-12
Identities = 36/104 (34%), Positives = 61/104 (58%)
Frame = +2
Query: 371 NAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAF 430
N +A++ L LY+ + G + W++ +EI+PLR RG IA + +N +V+ +F
Sbjct: 1478 NFGWIAILALALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSF 1657
Query: 431 SPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
L +G F IF+ IA+ ++ F+ VPETKG+ +EE+E+
Sbjct: 1658 LSLTVAIGPAWTFMIFAIIAIVAIFFVIIFVPETKGVPMEEVES 1789
>TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (55%)
Length = 949
Score = 122 bits (306), Expect = 3e-28
Identities = 85/271 (31%), Positives = 131/271 (47%), Gaps = 10/271 (3%)
Frame = +1
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIAD 87
GGLLFGYD G S A + I+ L + + +G++ GA G L D
Sbjct: 199 GGLLFGYDTGVISGALLYIKDDFPQVRNSNFLQETIVSMAIAGAIVGAAFGGWLN----D 366
Query: 88 FLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTP 147
GR++ L+A +++++GA++ A AP+ +L+ GRL+ G+G+G+A AP+YIAE AP+
Sbjct: 367 AYGRKKATLLADVIFILGAILMAAAPDPYVLIAGRLLVGLGVGIASVTAPVYIAEVAPSE 546
Query: 148 IRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
IRG LVS I G Y + + WR+M G+S A+I M +LP SPR
Sbjct: 547 IRGSLVSTNVLMITGGQFVSYLVNLVFTQVPGTWRWMLGVSGVPALIQFICMLFLPESPR 726
Query: 208 WILLR--------AIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGE 259
W+ ++ I K DL L+D ++D + A+ +
Sbjct: 727 WLFIKNRKNEAVDVISKIYDLSRLED--------------------EIDFLTAQSEQERQ 846
Query: 260 -ENDVTLGEMFRGK-CRKALVISAGLVLFQQ 288
+ + +FR K R A + GL+ FQQ
Sbjct: 847 RRSTIKFWHVFRSKETRLAFLDGGGLLAFQQ 939
>BF519718 weakly similar to PIR|G84564|G84 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (20%)
Length = 659
Score = 115 bits (287), Expect = 5e-26
Identities = 61/160 (38%), Positives = 95/160 (59%), Gaps = 7/160 (4%)
Frame = +3
Query: 322 ILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN-------AAV 374
I++G+ K + +V+DR GRRP+LL G SG+ +SLF LG L N A
Sbjct: 3 IIMGIAKTCFVLFSALVLDRFGRRPMLLLGSSGMAVSLFGLGMGCTLLHNSDEKPMWAIA 182
Query: 375 LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLK 434
L VV + V + I GP W+ +EIFP+RLR +G S+A+ VN + +V+ +F +
Sbjct: 183 LCVVAVCAAVSFFSIGLGPTTWVYSSEIFPMRLRAQGTSLAISVNRLISGVVSMSFLSIS 362
Query: 435 DLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
+ + G +F++ + + V + +F Y+ +PETKG +LEEIEA
Sbjct: 363 EEITFGGMFFVLAGVMVLATLFFYYFLPETKGKSLEEIEA 482
>BI266011 similar to GP|9759246|dbj| sugar transporter-like protein
{Arabidopsis thaliana}, partial (15%)
Length = 533
Score = 114 bits (284), Expect = 1e-25
Identities = 53/79 (67%), Positives = 68/79 (85%)
Frame = +3
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L F+FPA GGLLFGYDIGATS ATIS+QS LSG+ W +L ++++GL+ SGSLYGAL+GS
Sbjct: 297 LPFVFPALGGLLFGYDIGATSGATISLQSPELSGVAWSNLSSIQLGLVVSGSLYGALLGS 476
Query: 80 VLAFNIADFLGRRRELLVA 98
+LAF IADF+GR+R+L+ A
Sbjct: 477 LLAFAIADFIGRKRQLIGA 533
>TC81269 similar to GP|23504385|emb|CAC00697. putative sugar transporter
{Lycopersicon esculentum}, partial (42%)
Length = 966
Score = 113 bits (283), Expect = 1e-25
Identities = 69/206 (33%), Positives = 121/206 (58%), Gaps = 6/206 (2%)
Frame = +2
Query: 275 KALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGV 334
++L I GL++ QQ+ + S+ + GF A SI+ + ++++TGV
Sbjct: 35 RSLTIGVGLMVCQQLGGINGVCFYTSSIF---DLAGFPSATG----SIIYAILQIVITGV 193
Query: 335 AVVVVDRLGRRPLLLGGVSGIVIS-LFLLGSYYIFLDNAAV-----LAVVGLLLYVGCYQ 388
++DR GR+PLLL SG+V +F ++Y+ + + AV LAV G+L+Y+G +
Sbjct: 194 GAALIDRAGRKPLLLVSGSGLVAGCIFTAVAFYLKVHDVAVGAVPALAVTGILVYIGSFS 373
Query: 389 ISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSA 448
I G + W++++EIFP+ ++G+ SIA LVN+ L ++ F+ L G F +++A
Sbjct: 374 IGMGAIPWVVMSEIFPVNIKGQAGSIATLVNWFGAWLCSYTFNFLMSWSSYG-TFVLYAA 550
Query: 449 IAVASLVFIYFIVPETKGLTLEEIEA 474
I +++FI +VPETKG +LE+++A
Sbjct: 551 INALAILFIAVVVPETKGKSLEQLQA 628
>TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar transporter
{Prunus armeniaca}, partial (59%)
Length = 1074
Score = 113 bits (283), Expect = 1e-25
Identities = 84/302 (27%), Positives = 155/302 (50%), Gaps = 6/302 (1%)
Frame = +1
Query: 178 VAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGR 237
++ WR MFGI+ ++++ GM P SPRW+ ++G + A +++ L G+
Sbjct: 10 LSAWRTMFGIAVVPSILLALGMAISPESPRWLF-----QQGKIAE----AEKAIKTLYGK 162
Query: 238 TFHDSAPQQVDEIMAEF---SYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTD 294
++V +M + S E + ++F + K + + A L LFQQ+ +
Sbjct: 163 -------ERVATVMYDLRTASQGSSEPEAGWFDLFSSRYWKVVSVGAALFLFQQLAGINA 321
Query: 295 HGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSG 354
S+ R +A+D S L+G +I T +A ++D+ GR+ LL+ SG
Sbjct: 322 VVYYSTSVFRSAG-----IASDVA-ASALVGASNVIGTAIASSLMDKQGRKSLLITSFSG 483
Query: 355 IVISLFLLG---SYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKG 411
+ S+ LL ++ + + LAV+G +LYV + + GP+ L++ EIF R+R K
Sbjct: 484 MAASMLLLSLSFTWKVLAPYSGTLAVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKA 663
Query: 412 LSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEE 471
+S+++ ++ +N ++ F + + G ++ FSA+ V ++++I V ETKG +LEE
Sbjct: 664 VSLSLGTHWISNFVIGLYFLSVVNKFGISSVYLGFSAVCVLAVLYIAGNVVETKGRSLEE 843
Query: 472 IE 473
IE
Sbjct: 844 IE 849
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 106 bits (264), Expect = 2e-23
Identities = 87/333 (26%), Positives = 151/333 (45%), Gaps = 2/333 (0%)
Frame = +3
Query: 31 LFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFLG 90
LFGY +G + S+S ++ + + GL+ S L GAL G +L+ IAD +G
Sbjct: 411 LFGYHLGVVNEPL-----ESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIADAVG 575
Query: 91 RRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRG 150
RRR + AL ++GA ++A N +++GRL G G+GL A +Y+ E +P +RG
Sbjct: 576 RRRAFQLCALPMIIGAAMSAATNNLFGMLVGRLFVGTGLGLGPPVAALYVTEVSPAFVRG 755
Query: 151 QLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWIL 210
+L + GI+ +G + + WR F +S+ A I+ M + SP W+
Sbjct: 756 TYGALIQIATCFGILGSLFIGIPVKEISGWWRVCFWVSTIPAAILALAMDFCAESPHWLY 935
Query: 211 LRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEND-VTLGEMF 269
+ + + + L + F S +VD GE+ D V E+
Sbjct: 936 KQGRTAEAEAE------FERLLGVSEAKFAMSQLSKVDR--------GEDTDTVKFSELL 1073
Query: 270 RGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGF-SLAADATRVSILLGVFK 328
G K + I + L QQ+ + + + + T F S + ++ +GV
Sbjct: 1074HGHHSKVVFIGSTLFALQQLSGI--------NAVFYFSSTVFKSAGVPSDFANVCIGVAN 1229
Query: 329 LIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFL 361
L + ++ ++D+LGR+ LL G+ IS+ +
Sbjct: 1230LTGSIISTGLMDKLGRKVLLFWSFFGMAISMII 1328
>TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90
{Arabidopsis thaliana}, partial (41%)
Length = 827
Score = 102 bits (254), Expect = 3e-22
Identities = 53/163 (32%), Positives = 97/163 (58%), Gaps = 6/163 (3%)
Frame = +2
Query: 321 SILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY------IFLDNAAV 374
+I ++ T + +++D+ GRRPL+ SG + F+ G + + L+ +
Sbjct: 122 TIAYACMQVPFTILGAILMDKSGRRPLITASASGTFLGCFMTGIAFFLKDQNLLLELVPI 301
Query: 375 LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLK 434
LAV G+L+YV + I GP+ W++++EIFP+ ++G S+ VL+N+ +V++ F+
Sbjct: 302 LAVAGILIYVAAFSIGMGPVPWVIMSEIFPIHVKGTAGSLVVLINWLGAWVVSYTFNFFL 481
Query: 435 DLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
G LF +++ ++ +++F+ +VPETKG TLEEI+A CL
Sbjct: 482 SWSSPGTLF-LYAGCSLLTILFVAKLVPETKGKTLEEIQA-CL 604
>TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (88%)
Length = 2467
Score = 100 bits (249), Expect = 1e-21
Identities = 66/203 (32%), Positives = 114/203 (55%), Gaps = 4/203 (1%)
Frame = +3
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
+ G L G+D AT + +I L+ L GL+ + SL GA + + + I
Sbjct: 144 SIGNFLQGWD-NATIAGSILYIKKDLA------LQTTMEGLVVAMSLIGATVITTCSGPI 302
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
+D+LGRR +++++++Y +G+L+ ++PN +L + RL+ G GIGLA+ P+YI+ETAP
Sbjct: 303 SDWLGRRPMMIISSVLYFLGSLVMLWSPNVYVLCLARLLDGFGIGLAVTLVPVYISETAP 482
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGLGSLL-VDTVAGWRYMFGI-SSPVAVIMGFGMWWLP 203
+ IRG L +L +F G+ Y + ++ + WR M G+ S P +++LP
Sbjct: 483 SDIRGSLNTLPQFSGSGGMFLSYCMVFVMSLSPSPSWRIMLGVLSIPSLFYFLLTVFFLP 662
Query: 204 ASPRWILLRA--IQKKGDLQTLK 224
SPRW++ + ++ K LQ L+
Sbjct: 663 ESPRWLVSKGKMLEAKKVLQRLR 731
Score = 87.4 bits (215), Expect = 1e-17
Identities = 59/208 (28%), Positives = 109/208 (52%), Gaps = 9/208 (4%)
Frame = +3
Query: 274 RKALVISAGLVLFQQIQTV------TDHGPTKCSLLRCINPTGFSLAADATRVSILLGVF 327
+ AL++ G+ L QQ + T + + + G S + + +S + +
Sbjct: 1626 KHALIVGIGIQLLQQFSGINGVLYYTPQILEEAGVAVLLADLGLSSTSSSFLISAVTTLL 1805
Query: 328 KLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLL--GSYYIFLDNA-AVLAVVGLLLYV 384
L G+A+ ++D GRR LLL + +++SL +L GS F A ++ V +++Y
Sbjct: 1806 MLPSIGLAMRLMDVTGRRQLLLVTIPVLIVSLVILVLGSVIDFGSVVHAAISTVCVVVYF 1985
Query: 385 GCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFY 444
+ + +GP+ ++ +EIFP R+RG ++I LV + + +VT++ + LG +F
Sbjct: 1986 CFFVMGYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIVTYSLPVMLSSLGLAGVFG 2165
Query: 445 IFSAIAVASLVFIYFIVPETKGLTLEEI 472
+++ + S VF+Y VPETKG+ LE I
Sbjct: 2166 VYAIVCCISWVFVYLKVPETKGMPLEVI 2249
Score = 28.1 bits (61), Expect = 7.7
Identities = 46/190 (24%), Positives = 76/190 (39%), Gaps = 42/190 (22%)
Frame = +3
Query: 47 QSSSLSGITWYDLDAVE----------IGLLTSGSLYGALIGSV----------LAFNIA 86
Q S ++G+ +Y +E +GL ++ S + LI +V LA +
Sbjct: 1668 QFSGINGVLYYTPQILEEAGVAVLLADLGLSSTSSSF--LISAVTTLLMLPSIGLAMRLM 1841
Query: 87 DFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAA---------- 136
D GRR+ LLV + +V +I LV+G + I G +HAA
Sbjct: 1842 DVTGRRQLLLVTIPVLIVSLVI---------LVLGSV---IDFGSVVHAAISTVCVVVYF 1985
Query: 137 -----------PMYIAETAPTPIRGQLVSLKEFFIVIG-IVAGYGLGSLLVDTVAGWRYM 184
+ +E PT +RG +++ IG I+ Y L +L + G +
Sbjct: 1986 CFFVMGYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIVTYSLPVML--SSLGLAGV 2159
Query: 185 FGISSPVAVI 194
FG+ + V I
Sbjct: 2160 FGVYAIVCCI 2189
>TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 664
Score = 99.4 bits (246), Expect = 3e-21
Identities = 58/165 (35%), Positives = 91/165 (55%), Gaps = 3/165 (1%)
Frame = +2
Query: 20 LRFLFPA-FGGLLFGYDIGATSSATISIQSS--SLSGITWYDLDAVEIGLLTSGSLYGAL 76
LR F A GG LFGYD G S A + I+ ++ TW + S +L GA+
Sbjct: 188 LRLAFSAGIGGFLFGYDTGVISGALLYIRDDFKAVDRQTWLQ------EAIVSTALAGAI 349
Query: 77 IGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAA 136
IG+ + I D GR++ +++A ++ +G++I A A N +L++GR+ G+G+G+A A+
Sbjct: 350 IGASVGGWINDRFGRKKAIILADALFFIGSVIMAAAINPAILIVGRVFVGLGVGMASMAS 529
Query: 137 PMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGW 181
P+YI+E +PT +RG LVSL F I G Y + + W
Sbjct: 530 PLYISEASPTRVRGALVSLNGFLITGGQXLSYVINLAFTNAPGTW 664
>TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (49%)
Length = 1353
Score = 96.3 bits (238), Expect = 2e-20
Identities = 60/187 (32%), Positives = 100/187 (53%), Gaps = 2/187 (1%)
Frame = +2
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
A G LL G+D + + + I+ VE GL+ + SL GA + + + +
Sbjct: 185 AIGNLLQGWDNATIAGSILYIKRE----FQLQSEPTVE-GLIVAMSLIGATVVTTCSGAL 349
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
+D GRR L++++L+Y + +L+ ++PN +L+ RL+ G+GIGLA+ P+YI+E AP
Sbjct: 350 SDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAP 529
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGL-GSLLVDTVAGWRYMFGI-SSPVAVIMGFGMWWLP 203
IRG L +L +F G+ Y + + + WR M G+ S P + + LP
Sbjct: 530 PEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFALTLLVLP 709
Query: 204 ASPRWIL 210
SPRW++
Sbjct: 710 ESPRWLV 730
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.145 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,404,646
Number of Sequences: 36976
Number of extensions: 205450
Number of successful extensions: 1299
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 1211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1237
length of query: 477
length of database: 9,014,727
effective HSP length: 100
effective length of query: 377
effective length of database: 5,317,127
effective search space: 2004556879
effective search space used: 2004556879
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142222.8


