

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.6 - phase: 0
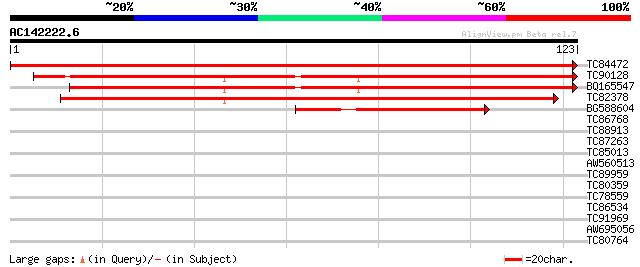
(123 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84472 similar to GP|7770334|gb|AAF69704.1| F27J15.21 {Arabidop... 244 5e-66
TC90128 similar to GP|7770334|gb|AAF69704.1| F27J15.21 {Arabidop... 145 4e-36
BQ165547 similar to GP|7770334|gb|A F27J15.21 {Arabidopsis thali... 142 3e-35
TC82378 weakly similar to GP|7770334|gb|AAF69704.1| F27J15.21 {A... 102 3e-23
BG588604 similar to PIR|G96739|G967 F14O23.12 [imported] - Arabi... 48 1e-06
TC86768 similar to GP|15294244|gb|AAK95299.1 At2g46600/F13A10.13... 28 0.68
TC88913 homologue to PIR|S55945|S55945 zinc proteinase (EC 3.4.2... 26 4.4
TC87263 similar to GP|9759298|dbj|BAB09804.1 emb|CAB82953.1~gene... 26 4.4
TC85013 similar to PIR|T50562|T50562 SINA2 protein [imported] - ... 26 4.4
AW560513 25 5.7
TC89959 similar to GP|15795137|dbj|BAB02515. transporter-like pr... 25 5.7
TC80359 GP|1435157|emb|CAA58445.1 histone H3 variant H3.3 {Lycop... 25 5.7
TC78559 weakly similar to GP|5733686|gb|AAD49719.1| maturation p... 25 5.7
TC86534 homologue to SP|P12227|RPOD_PEA DNA-directed RNA polymer... 25 7.5
TC91969 similar to GP|9758643|dbj|BAB09267.1 fimbrin {Arabidopsi... 25 9.8
AW695056 25 9.8
TC80764 similar to GP|12039376|gb|AAG46162.1 hypothetical protei... 25 9.8
>TC84472 similar to GP|7770334|gb|AAF69704.1| F27J15.21 {Arabidopsis
thaliana}, partial (25%)
Length = 751
Score = 244 bits (624), Expect = 5e-66
Identities = 123/123 (100%), Positives = 123/123 (100%)
Frame = +1
Query: 1 MANLTSSPESLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNMSNKALSQF 60
MANLTSSPESLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNMSNKALSQF
Sbjct: 67 MANLTSSPESLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNMSNKALSQF 246
Query: 61 GKMKKTKKQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYDINGRQTGEVPLRSPRAIPF 120
GKMKKTKKQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYDINGRQTGEVPLRSPRAIPF
Sbjct: 247 GKMKKTKKQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYDINGRQTGEVPLRSPRAIPF 426
Query: 121 SRY 123
SRY
Sbjct: 427 SRY 435
>TC90128 similar to GP|7770334|gb|AAF69704.1| F27J15.21 {Arabidopsis
thaliana}, partial (32%)
Length = 716
Score = 145 bits (366), Expect = 4e-36
Identities = 85/147 (57%), Positives = 100/147 (67%), Gaps = 29/147 (19%)
Frame = +3
Query: 6 SSPESLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKT-------------------- 45
+SPES + + SN FF+TISAILA+LSR+A RLK KAK+
Sbjct: 57 TSPESPN-RHHSNGGFFVTISAILALLSRKANRLKEKAKSSSTTKPIRDEEWRFDLKTPP 233
Query: 46 -------KKLLSNMSNKALSQFGKMKKTKKQRDNEL--NDGVWQKEILMGGKCEPLDFSG 96
KKLLSN+SNKALSQFGK KK +++R+ E N GVWQKEILMGGKCEPLDFSG
Sbjct: 234 KSPMAKPKKLLSNISNKALSQFGK-KKQREEREKEGWGNGGVWQKEILMGGKCEPLDFSG 410
Query: 97 VIHYDINGRQTGEVPLRSPRAIPFSRY 123
VI+YDING+QT EVP+RSPRA P Y
Sbjct: 411 VIYYDINGKQTREVPIRSPRASPLPGY 491
>BQ165547 similar to GP|7770334|gb|A F27J15.21 {Arabidopsis thaliana},
partial (32%)
Length = 649
Score = 142 bits (358), Expect = 3e-35
Identities = 81/139 (58%), Positives = 94/139 (67%), Gaps = 29/139 (20%)
Frame = -1
Query: 14 KRQSNSSFFITISAILAMLSRQARRLKGKAKT---------------------------K 46
+ SN FF+TISAILA+LSR+A RLK KAK+ K
Sbjct: 646 RHHSNGGFFVTISAILALLSRKASRLKEKAKSSSTTKPIRDEEWRFDLKTPPKSPMAKPK 467
Query: 47 KLLSNMSNKALSQFGKMKKTKKQRDNEL--NDGVWQKEILMGGKCEPLDFSGVIHYDING 104
KLLSN+SNKALSQFGK KK +++R+ E N GVWQKEILMGGKCEPLDFSGVI+YDING
Sbjct: 466 KLLSNISNKALSQFGK-KKQREEREKEGWGNGGVWQKEILMGGKCEPLDFSGVIYYDING 290
Query: 105 RQTGEVPLRSPRAIPFSRY 123
+QT EVP+RSPRA P Y
Sbjct: 289 KQTREVPIRSPRASPLPGY 233
>TC82378 weakly similar to GP|7770334|gb|AAF69704.1| F27J15.21 {Arabidopsis
thaliana}, partial (37%)
Length = 568
Score = 102 bits (255), Expect = 3e-23
Identities = 57/114 (50%), Positives = 69/114 (60%), Gaps = 6/114 (5%)
Frame = +2
Query: 12 SPKRQSNSSFFITISAILAMLSRQARRLKGKAKT------KKLLSNMSNKALSQFGKMKK 65
S KR++N + + M+ R +LK T KKLL N+SNKA+ KMKK
Sbjct: 26 SSKRRANRFIINVTTLMTVMVKRATLKLKAAPMTSEVKSPKKLLKNISNKAMPFIEKMKK 205
Query: 66 TKKQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYDINGRQTGEVPLRSPRAIP 119
KK + + GVWQK ILMG KCEPLDFSGVI+YD G+Q E PLRSPRA P
Sbjct: 206 KKKGKLEWGDGGVWQKTILMGDKCEPLDFSGVIYYDNKGKQVNEFPLRSPRATP 367
>BG588604 similar to PIR|G96739|G967 F14O23.12 [imported] - Arabidopsis
thaliana, partial (32%)
Length = 568
Score = 47.8 bits (112), Expect = 1e-06
Identities = 23/42 (54%), Positives = 29/42 (68%)
Frame = +3
Query: 63 MKKTKKQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYDING 104
M ++QR+ D +WQK ILMGGKC+ DFSGVI YD +G
Sbjct: 156 MNGCEQQREE---DSIWQKNILMGGKCQLPDFSGVIIYDSDG 272
>TC86768 similar to GP|15294244|gb|AAK95299.1 At2g46600/F13A10.13
{Arabidopsis thaliana}, partial (85%)
Length = 651
Score = 28.5 bits (62), Expect = 0.68
Identities = 16/54 (29%), Positives = 27/54 (49%)
Frame = -1
Query: 4 LTSSPESLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNMSNKAL 57
+ S+ L P S + FF+ +S ++ +S ARRLK + +S S A+
Sbjct: 351 IASASSLLIPSMPSMAEFFLRLSLVIKPVSWSARRLKPPHSSDTKVSTSSFSAI 190
>TC88913 homologue to PIR|S55945|S55945 zinc proteinase (EC 3.4.24.-) STE23 -
yeast (Saccharomyces cerevisiae), partial (1%)
Length = 1735
Score = 25.8 bits (55), Expect = 4.4
Identities = 12/40 (30%), Positives = 20/40 (50%)
Frame = -3
Query: 51 NMSNKALSQFGKMKKTKKQRDNELNDGVWQKEILMGGKCE 90
++ NK SQ T K+ + +N W++ +GG CE
Sbjct: 1040 SLKNKCQSQ------TSKEG*SSINSQAWRRRKFLGGHCE 939
>TC87263 similar to GP|9759298|dbj|BAB09804.1
emb|CAB82953.1~gene_id:MPH15.5~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (65%)
Length = 2229
Score = 25.8 bits (55), Expect = 4.4
Identities = 13/32 (40%), Positives = 20/32 (61%)
Frame = +3
Query: 23 ITISAILAMLSRQARRLKGKAKTKKLLSNMSN 54
I++S+ILA + LKG+ TKK+ +M N
Sbjct: 1317 ISLSSILATGGLMIKLLKGRTITKKVAMSMMN 1412
>TC85013 similar to PIR|T50562|T50562 SINA2 protein [imported] - Vitis
vinifera, partial (25%)
Length = 666
Score = 25.8 bits (55), Expect = 4.4
Identities = 9/33 (27%), Positives = 19/33 (57%)
Frame = +3
Query: 62 KMKKTKKQRDNELNDGVWQKEILMGGKCEPLDF 94
K+KK +++ + + +G W++ + PLDF
Sbjct: 255 KLKKLERKPLDRIRNGSWRRYFQRSSRVSPLDF 353
>AW560513
Length = 416
Score = 25.4 bits (54), Expect = 5.7
Identities = 11/24 (45%), Positives = 17/24 (70%)
Frame = -3
Query: 63 MKKTKKQRDNELNDGVWQKEILMG 86
+KKTK ++ + NDGV K++L G
Sbjct: 384 IKKTKLSKEEKQNDGVKLKKVL*G 313
>TC89959 similar to GP|15795137|dbj|BAB02515. transporter-like protein
{Arabidopsis thaliana}, partial (87%)
Length = 1681
Score = 25.4 bits (54), Expect = 5.7
Identities = 18/59 (30%), Positives = 28/59 (46%), Gaps = 1/59 (1%)
Frame = -2
Query: 11 LSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSN-MSNKALSQFGKMKKTKK 68
LSPKRQ F+++ IL L + +GK K++ +SN L+ K +K
Sbjct: 783 LSPKRQLLQLFYLSSIGILVTLVSFCKTEEGKMWEGKIVQEAISNPVLASQSKQVMLQK 607
>TC80359 GP|1435157|emb|CAA58445.1 histone H3 variant H3.3 {Lycopersicon
esculentum}, complete
Length = 1440
Score = 25.4 bits (54), Expect = 5.7
Identities = 17/53 (32%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Frame = +2
Query: 19 SSFFITISAILAMLSRQARRLKG-KAKTKKLLSNMSNKALSQFGKMKKTKKQR 70
SS F S +A + AR+ G KA K+L + + K+ G +KK + R
Sbjct: 23 SSSFFRFSNSMARTKQTARKSTGGKAPRKQLATKAARKSAPTTGGVKKPHRYR 181
>TC78559 weakly similar to GP|5733686|gb|AAD49719.1| maturation protein
pPM32 {Glycine max}, partial (55%)
Length = 1128
Score = 25.4 bits (54), Expect = 5.7
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = -1
Query: 5 TSSPESLSPKRQSNSSFFITISAILAMLS 33
T++PE P+ S+SSF T+S I + S
Sbjct: 837 TTTPEPPLPRSSSSSSFPTTVSLIFCVAS 751
>TC86534 homologue to SP|P12227|RPOD_PEA DNA-directed RNA polymerase beta'
chain (EC 2.7.7.6) (Fragment). [Garden pea] {Pisum
sativum}, partial (61%)
Length = 2441
Score = 25.0 bits (53), Expect = 7.5
Identities = 9/21 (42%), Positives = 14/21 (65%)
Frame = -1
Query: 80 QKEILMGGKCEPLDFSGVIHY 100
Q +IL G C+PLD +I++
Sbjct: 1988 QNKILFLGSCDPLDIDPIINF 1926
>TC91969 similar to GP|9758643|dbj|BAB09267.1 fimbrin {Arabidopsis
thaliana}, partial (36%)
Length = 781
Score = 24.6 bits (52), Expect = 9.8
Identities = 14/53 (26%), Positives = 29/53 (54%)
Frame = -3
Query: 3 NLTSSPESLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNMSNK 55
+++S+ E+++P S +SFF I++ + ++ K +L N+SNK
Sbjct: 635 DISSNSETVNP--DSEASFFPRCLCIISHHLSEGHFIRVNCKPIPMLENLSNK 483
>AW695056
Length = 515
Score = 24.6 bits (52), Expect = 9.8
Identities = 11/30 (36%), Positives = 17/30 (56%)
Frame = -1
Query: 50 SNMSNKALSQFGKMKKTKKQRDNELNDGVW 79
SN++ ++L+Q K +R LND VW
Sbjct: 170 SNLAKRSLNQNAKRHMQTHKRCPSLNDQVW 81
>TC80764 similar to GP|12039376|gb|AAG46162.1 hypothetical protein {Oryza
sativa}, partial (25%)
Length = 928
Score = 24.6 bits (52), Expect = 9.8
Identities = 10/20 (50%), Positives = 15/20 (75%)
Frame = +2
Query: 5 TSSPESLSPKRQSNSSFFIT 24
TSSPESL+P+ S+S ++
Sbjct: 110 TSSPESLAPEAHSSSHLLLS 169
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,010,702
Number of Sequences: 36976
Number of extensions: 30598
Number of successful extensions: 207
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 203
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 203
length of query: 123
length of database: 9,014,727
effective HSP length: 84
effective length of query: 39
effective length of database: 5,908,743
effective search space: 230440977
effective search space used: 230440977
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC142222.6


