

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.5 + phase: 0
(378 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
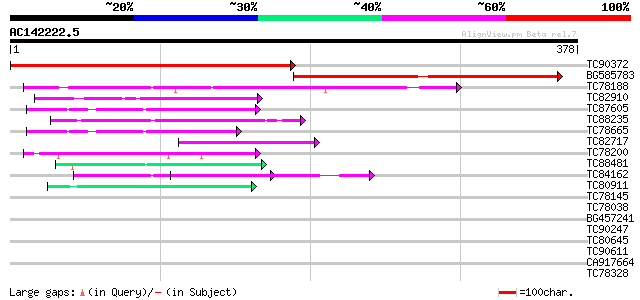
Score E
Sequences producing significant alignments: (bits) Value
TC90372 similar to GP|20197166|gb|AAC27402.2 expressed protein {... 373 e-104
BG585783 similar to GP|20197166|gb expressed protein {Arabidopsi... 311 3e-85
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 72 4e-13
TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Ar... 59 2e-09
TC87605 homologue to PIR|G96563|G96563 probable coatomer complex... 52 4e-07
TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Or... 50 2e-06
TC78665 homologue to PIR|G96563|G96563 probable coatomer complex... 49 3e-06
TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coa... 44 1e-04
TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat prote... 44 1e-04
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 42 3e-04
TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repea... 42 3e-04
TC80911 similar to GP|3668118|emb|CAA11819.1 hypothetical protei... 41 7e-04
TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {... 40 0.001
TC78038 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Ara... 40 0.001
BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [impo... 40 0.002
TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Ara... 39 0.003
TC80645 similar to PIR|T50983|T50983 probable pleiotropic regula... 39 0.003
TC90611 similar to GP|10177223|dbj|BAB10298. contains similarity... 39 0.004
CA917664 similar to GP|16604679|gb putative WD-repeat membrane p... 38 0.007
TC78328 similar to GP|11994333|dbj|BAB02292. WD-40 repeat protei... 37 0.010
>TC90372 similar to GP|20197166|gb|AAC27402.2 expressed protein {Arabidopsis
thaliana}, partial (52%)
Length = 706
Score = 373 bits (958), Expect = e-104
Identities = 183/190 (96%), Positives = 190/190 (99%)
Frame = +1
Query: 1 MEINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAA 60
MEINLGKLAFDIDFHPS+NLVA+GLIDGDLHLYRYSSDNTNSDPVR+LE+HAHTESCRAA
Sbjct: 88 MEINLGKLAFDIDFHPSENLVASGLIDGDLHLYRYSSDNTNSDPVRVLEVHAHTESCRAA 267
Query: 61 RFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCI 120
RFINGGRA+LTGSPDFSILATDVETGSTIARLDNAHEAAVNRL+NLTESTVASGDDDGCI
Sbjct: 268 RFINGGRAVLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLINLTESTVASGDDDGCI 447
Query: 121 KVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSE 180
KVWDTRERSCCNSFEAHEDYISDITFASDAMK+LATSGDGTLSVCNLRRNKVQAQSEFSE
Sbjct: 448 KVWDTRERSCCNSFEAHEDYISDITFASDAMKILATSGDGTLSVCNLRRNKVQAQSEFSE 627
Query: 181 DELLSVVLMK 190
DELLSVVLMK
Sbjct: 628 DELLSVVLMK 657
>BG585783 similar to GP|20197166|gb expressed protein {Arabidopsis thaliana},
partial (32%)
Length = 743
Score = 311 bits (796), Expect = 3e-85
Identities = 152/179 (84%), Positives = 164/179 (90%)
Frame = +3
Query: 190 KNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINL 249
+NGRKVVCGSQTGI+LLYSWGCFKDCSDRFVDL+SNSIDTMLKLDEDRIITGSENGMINL
Sbjct: 36 QNGRKVVCGSQTGILLLYSWGCFKDCSDRFVDLASNSIDTMLKLDEDRIITGSENGMINL 215
Query: 250 VGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNILQGSR 309
VGILPNR+I+P+AEHSEYPVERL AFSHDRKFLGSI HDQMLKLWDLDNILQGSR
Sbjct: 216 VGILPNRIIEPIAEHSEYPVERL------AFSHDRKFLGSIGHDQMLKLWDLDNILQGSR 377
Query: 310 NTQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKRKNASKGHAAGDSNNFFADL*EI 368
+TQRNE G V ND DSD+ DEMDVDN+ SKF+KGNKRKNAS GHA GDSNNFFADL*++
Sbjct: 378 STQRNETGVVANDGDSDDGDEMDVDNSASKFSKGNKRKNASNGHAVGDSNNFFADL*DM 554
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 72.0 bits (175), Expect = 4e-13
Identities = 60/301 (19%), Positives = 128/301 (41%), Gaps = 9/301 (2%)
Frame = +1
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + F+P+ ++VA+G D ++ L+ N + D + + H + + + G +
Sbjct: 325 YTMKFNPTGSVVASGSHDKEIFLW-----NVHGDCKNFMVLKGHKNAVLDLHWTSDGTQI 489
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES--TVASGDDDGCIKVWDTRE 127
++ SPD ++ D ETG I ++ H + VN V SG DDG K+WD R+
Sbjct: 490 ISASPDKTLRLWDTETGKQIKKMVE-HLSYVNSCCPTRRGPPLVVSGSDDGTAKLWDMRQ 666
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV 187
R +F + I+ ++F+ + K+ D + + +LR+ +V + +D + S+
Sbjct: 667 RGSIQTF-PDKYQITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQGHQDMITSMQ 843
Query: 188 LMKNGRKVVCGSQTGIMLLYSW-------GCFKDCSDRFVDLSSNSIDTMLKLDEDRIIT 240
L +G ++ + ++ C K + N + D ++
Sbjct: 844 LSPDGSYLLTNGMDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCGWSPDGSKVTA 1023
Query: 241 GSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
GS + M+ + R++ + H+ G+ F + +GS + D+ + L +
Sbjct: 1024GSSDRMVYIWDTTSRRILYKLPGHN-------GSVNECVFHPNEPIVGSCSSDKQIYLGE 1182
Query: 301 L 301
+
Sbjct: 1183I 1185
Score = 38.9 bits (89), Expect = 0.003
Identities = 48/214 (22%), Positives = 87/214 (40%), Gaps = 8/214 (3%)
Frame = +1
Query: 96 HEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSCCNSF---EAHEDYISDITFASDAM 151
H++AV + N T S VASG D I +W+ C +F + H++ + D+ + SD
Sbjct: 310 HQSAVYTMKFNPTGSVVASGSHDKEIFLWNV--HGDCKNFMVLKGHKNAVLDLHWTSDGT 483
Query: 152 KLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRK-VVCGSQTGIMLLYSW- 209
++++ S D TL + + K + + S + G VV GS G L+
Sbjct: 484 QIISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPTRRGPPLVVSGSDDGTAKLWDMR 663
Query: 210 --GCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEY 267
G + D++ + + D D+I TG + + + + V + H +
Sbjct: 664 QRGSIQTFPDKYQITAVSFSDA-----SDKIYTGGIDNDVKIWDLRKGEVTMTLQGHQDM 828
Query: 268 PVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
+ S D +L + D L +WD+
Sbjct: 829 ITS-------MQLSPDGSYLLTNGMDCKLCIWDM 909
>TC82910 similar to GP|19347844|gb|AAL86002.1 unknown protein {Arabidopsis
thaliana}, partial (19%)
Length = 821
Score = 59.3 bits (142), Expect = 2e-09
Identities = 39/152 (25%), Positives = 70/152 (45%)
Frame = +1
Query: 17 SDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDF 76
+ + TG +DG +H+ ++ S + +H+ S F G G D
Sbjct: 76 TSTIALTGSVDGSVHIVNITTGRVVST------LPSHSSSIECVGFAPSGSWAAIGGMD- 234
Query: 77 SILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEA 136
+ DVE ++AR HE V L S VA+G +DG +++WD+R C +F
Sbjct: 235 KMTIWDVE--HSLARSICEHEYGVTCSTWLGTSYVATGSNDGAVRLWDSRSGECVRTFRG 408
Query: 137 HEDYISDITFASDAMKLLATSGDGTLSVCNLR 168
H + I ++ +++ L++ S D T V +++
Sbjct: 409 HSESIQSLSLSANQEYLVSASLDHTARVFDVK 504
>TC87605 homologue to PIR|G96563|G96563 probable coatomer complex subunit
33791-27676 [imported] - Arabidopsis thaliana, partial
(23%)
Length = 765
Score = 52.0 bits (123), Expect = 4e-07
Identities = 41/159 (25%), Positives = 72/159 (44%), Gaps = 3/159 (1%)
Frame = +3
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+D HP++ + L G + ++ Y S T + + E+ R+A+F+ + ++
Sbjct: 186 VDLHPTEPWILASLYSGTVCIWNYQSQ-TMAKSFEVTELPV-----RSAKFVARKQWVVA 347
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSC 130
G+ D I + T + ++ AH + + V+ T V S DD IK+WD +
Sbjct: 348 GADDMFIRVYNYNTMDKV-KVFEAHTDYIRCVAVHPTLPYVLSSSDDMLIKLWDWEKGWI 524
Query: 131 CNS-FEAHEDYISDITF-ASDAMKLLATSGDGTLSVCNL 167
C FE H Y+ +TF D + S D T+ + NL
Sbjct: 525 CTQIFEGHSHYVMQVTFNPKDTNTFASASLDRTIKIWNL 641
>TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Oryza
sativa}, partial (91%)
Length = 1080
Score = 49.7 bits (117), Expect = 2e-06
Identities = 44/172 (25%), Positives = 76/172 (43%), Gaps = 2/172 (1%)
Frame = +2
Query: 28 GDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGS 87
G+ H+ + ++ + PV + HT + A F G + +GS D ++ D+
Sbjct: 371 GNPHIRLFDVNSNSPQPV--MSYDGHTSNVMAVGFQCDGNWMYSGSEDGTVKIWDLRAPG 544
Query: 88 TIARLDNAHEAAVNRLV-NLTESTVASGDDDGCIKVWDTRERSC-CNSFEAHEDYISDIT 145
++ AAVN +V + ++ + SGD +G I+VWD SC C + + +T
Sbjct: 545 CQREYES--RAAVNTVVLHPNQTELISGDQNGNIRVWDLTANSCSCELVPEVDTAVRSLT 718
Query: 146 FASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVC 197
D ++A + +GT V L R Q + F E L + NG + C
Sbjct: 719 VMWDGSLVVAANNNGTCYVWRLLRG-TQTMTNF---EPLHKLQAHNGYILKC 862
Score = 38.9 bits (89), Expect = 0.003
Identities = 24/98 (24%), Positives = 45/98 (45%), Gaps = 1/98 (1%)
Frame = +2
Query: 113 SGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKV 172
SG +DG +K+WD R C +E+ ++ + + +L++ +G + V +L N
Sbjct: 494 SGSEDGTVKIWDLRAPGCQREYESRA-AVNTVVLHPNQTELISGDQNGNIRVWDLTANSC 670
Query: 173 QAQSEFSEDELL-SVVLMKNGRKVVCGSQTGIMLLYSW 209
+ D + S+ +M +G VV + G Y W
Sbjct: 671 SCELVPEVDTAVRSLTVMWDGSLVVAANNNG--TCYVW 778
>TC78665 homologue to PIR|G96563|G96563 probable coatomer complex subunit
33791-27676 [imported] - Arabidopsis thaliana, partial
(21%)
Length = 715
Score = 48.9 bits (115), Expect = 3e-06
Identities = 38/145 (26%), Positives = 65/145 (44%), Gaps = 2/145 (1%)
Frame = +2
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+D HP++ + L G + ++ Y S T + + E+ R+A+FI + ++
Sbjct: 185 VDLHPTEPWILASLYSGTVCIWNYQSQ-TMAKSFEVTELPV-----RSAKFIARKQWVVA 346
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSC 130
G+ D I + T + ++ AH + + V+ T V S DD IK+WD +
Sbjct: 347 GADDMFIRVYNYNTMDKV-KVFEAHSDYIRCVAVHPTLPYVLSSSDDMLIKLWDWEKGWI 523
Query: 131 CNS-FEAHEDYISDITFASDAMKLL 154
C FE H Y+ +TF K L
Sbjct: 524 CTQIFEGHSHYVMQVTFNPKRYKYL 598
Score = 29.3 bits (64), Expect = 2.6
Identities = 16/49 (32%), Positives = 24/49 (48%)
Frame = +2
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGD 159
V +G DD I+V++ FEAH DYI + +L++S D
Sbjct: 338 VVAGADDMFIRVYNYNTMDKVKVFEAHSDYIRCVAVHPTLPYVLSSSDD 484
>TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coatomer
complex subunit {Arabidopsis thaliana}, partial (29%)
Length = 1118
Score = 43.9 bits (102), Expect = 1e-04
Identities = 21/94 (22%), Positives = 45/94 (47%)
Frame = +1
Query: 113 SGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKV 172
SG DD KVWD ++C + E H++ ++ I + ++ S D T+ + + ++
Sbjct: 700 SGSDDYTAKVWDYDSKNCVQTLEGHKNNVTAICAHPEIPIIITASEDSTVKIWDAVTYRL 879
Query: 173 QAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLL 206
Q +F + + S+ K ++ G G +++
Sbjct: 880 QNTLDFGLERVWSIGYKKGSSQLAFGCDKGFIIV 981
Score = 29.3 bits (64), Expect = 2.6
Identities = 27/133 (20%), Positives = 48/133 (35%)
Frame = +3
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+D HP++ V GL G + ++ Y + T +++ E RAA+FI
Sbjct: 150 VDLHPTEPWVLLGLYSGTISIWNYQT-KTEEKSLKVSE-----SPVRAAKFI-------- 287
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCC 131
+ E+ + + DD I+V++ +
Sbjct: 288 ----------------------------------VRENWIIAATDDKYIRVYNYEKMEKI 365
Query: 132 NSFEAHEDYISDI 144
FEAH+DYI +
Sbjct: 366 IEFEAHKDYIRSL 404
Score = 28.1 bits (61), Expect = 5.8
Identities = 18/58 (31%), Positives = 29/58 (49%), Gaps = 2/58 (3%)
Frame = +1
Query: 109 STVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITF--ASDAMKLLATSGDGTLSV 164
ST AS DG +K+W + +FE H ++ + + ++D LL+ S D T V
Sbjct: 556 STFASASLDGTLKIWTIDSSAPNFTFEGHLKGMNCVDYFESNDKQYLLSGSDDYTAKV 729
>TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana, partial (80%)
Length = 1824
Score = 43.5 bits (101), Expect = 1e-04
Identities = 47/168 (27%), Positives = 73/168 (42%), Gaps = 10/168 (5%)
Frame = +1
Query: 10 FDIDFHPSDNLVATGLIDGDLH--LYRYSSDNTNSDPVRLLEIHAHTESCRAARFI-NGG 66
+ ID+ P LV L+ GD + +Y + + + + HT+S ++
Sbjct: 718 YAIDWSP---LVPGRLVSGDCNNSIYLWEPTSAATWNIEKTPFTGHTDSVEDLQWSPTEP 888
Query: 67 RALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLV--NLTESTVASGDDDGCIKVWD 124
+ S D SI D G + A AH+A VN L L +ASG DDG I + D
Sbjct: 889 HVFASCSVDKSIAIWDTRLGRSPAASFQAHKADVNVLSWNRLASCMLASGSDDGTISIRD 1068
Query: 125 TR----ERSCCNSFEAHEDYISDITFA-SDAMKLLATSGDGTLSVCNL 167
R S FE H+ I+ I ++ +A L +S D L++ +L
Sbjct: 1069LRLLKEGDSVVAHFEYHKHPITSIEWSPHEASSLAVSSADNQLTIWDL 1212
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 42.4 bits (98), Expect = 3e-04
Identities = 42/146 (28%), Positives = 56/146 (37%), Gaps = 5/146 (3%)
Frame = +1
Query: 31 HLYRYSSDNT----NSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETG 86
HL S+D T N L H E F G+ L T S D + DVET
Sbjct: 1120 HLATASADRTAKYWNDQGALLGTFKGHLERLARIAFHPSGKYLGTASYDKTWRLWDVETE 1299
Query: 87 STIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDIT 145
+ L H +V L + S AS D +VWD R + E H I I+
Sbjct: 1300 EELL-LQEGHSRSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVLALEGHVKSILGIS 1476
Query: 146 FASDAMKLLATSGDGTLSVCNLRRNK 171
F+ + L D T + +LR+ K
Sbjct: 1477 FSPNGYHLATGGEDNTCRIWDLRKKK 1554
Score = 34.3 bits (77), Expect = 0.081
Identities = 29/157 (18%), Positives = 63/157 (39%), Gaps = 2/157 (1%)
Frame = +1
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ +DFH +L A+ +D ++ + + +L + H +S F G L
Sbjct: 1339 YGLDFHHDGSLAASCGLDALARVWDLRTGRS------VLALEGHVKSILGISFSPNGYHL 1500
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST--VASGDDDGCIKVWDTRE 127
TG D + D+ ++ + AH ++++ + + + D KVW +R+
Sbjct: 1501 ATGGEDNTCRIWDLRKKKSLYTIP-AHSNLISQVKFEPQEGYFLVTASYDMTAKVWSSRD 1677
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
+ HE ++ + D ++ S D T+ +
Sbjct: 1678 FKPVKTLSGHEAKVTSLDVLGDGGYIVTVSHDRTIKL 1788
Score = 27.7 bits (60), Expect = 7.6
Identities = 12/34 (35%), Positives = 22/34 (64%)
Frame = +1
Query: 269 VERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLD 302
+ERL +AF K+LG+ ++D+ +LWD++
Sbjct: 1201 LERLAR---IAFHPSGKYLGTASYDKTWRLWDVE 1293
>TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repeat protein
{Arabidopsis thaliana}, partial (29%)
Length = 628
Score = 42.4 bits (98), Expect = 3e-04
Identities = 33/136 (24%), Positives = 59/136 (43%), Gaps = 1/136 (0%)
Frame = +1
Query: 43 DPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNR 102
D V ++ H + F + ++T S D +I + GS + + H ++V R
Sbjct: 79 DLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAISDGSCLKTFEG-HTSSVLR 255
Query: 103 LVNLTEST-VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGT 161
+ +T T + S DG +K+W + C +++ HED + + + LAT G
Sbjct: 256 ALFVTRGTQIVSCGADGLVKLWTVKSNECVATYDNHEDKVWALAVGRKT-ETLATGGSD- 429
Query: 162 LSVCNLRRNKVQAQSE 177
+V NL + A E
Sbjct: 430 -AVVNLWLDSTAADKE 474
Score = 42.0 bits (97), Expect = 4e-04
Identities = 22/136 (16%), Positives = 59/136 (43%)
Frame = +1
Query: 108 ESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNL 167
+ V + D I++W + SC +FE H + F + ++++ DG + + +
Sbjct: 148 DQCVVTASGDKTIRIWAISDGSCLKTFEGHTSSVLRALFVTRGTQIVSCGADGLVKLWTV 327
Query: 168 RRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSI 227
+ N+ A + ED++ ++ + + + G ++ L+ +D ++
Sbjct: 328 KSNECVATYDNHEDKVWALAVGRKTETLATGGSDAVVNLW------------LDSTAADK 471
Query: 228 DTMLKLDEDRIITGSE 243
+ + +E+ ++ G E
Sbjct: 472 EEAFRKEEEGVLKGQE 519
Score = 31.6 bits (70), Expect = 0.53
Identities = 31/145 (21%), Positives = 56/145 (38%), Gaps = 4/145 (2%)
Frame = +1
Query: 109 STVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLR 168
S V SG D VW + F+ H+ I + F+ ++ SGD T+ + +
Sbjct: 25 SLVCSGSQDRTACVWRLPDLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAIS 204
Query: 169 RNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSID 228
E +L + + G ++V G++ L++ S+ V N D
Sbjct: 205 DGSCLKTFEGHTSSVLRALFVTRGTQIVSCGADGLVKLWTVK-----SNECVATYDNHED 369
Query: 229 TMLKLDEDR----IITGSENGMINL 249
+ L R + TG + ++NL
Sbjct: 370 KVWALAVGRKTETLATGGSDAVVNL 444
>TC80911 similar to GP|3668118|emb|CAA11819.1 hypothetical protein {Brassica
napus}, partial (22%)
Length = 860
Score = 41.2 bits (95), Expect = 7e-04
Identities = 29/139 (20%), Positives = 55/139 (38%)
Frame = +2
Query: 26 IDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVET 85
++ D++L+ S+ +T L+ + + + G L G + D
Sbjct: 335 LENDVYLWNASNKST----AELVSVDEEDGPVTSVSWCPDGSRLAIGLDSSLVQVWDTIA 502
Query: 86 GSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDIT 145
+ L + H A V+ L + +G +G I D R RS NS+ H D + +
Sbjct: 503 NKQLTTLKSGHRAGVSSLAWNNSHILTTGGMNGKIVNNDVRVRSHINSYRGHTDEVCGLK 682
Query: 146 FASDAMKLLATSGDGTLSV 164
++ D KL + D + +
Sbjct: 683 WSLDGKKLASGGSDNVVHI 739
Score = 27.7 bits (60), Expect = 7.6
Identities = 23/92 (25%), Positives = 35/92 (38%)
Frame = +2
Query: 64 NGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVW 123
N L TG + I+ DV S I + +L +ASG D + +W
Sbjct: 563 NNSHILTTGGMNGKIVNNDVRVRSHINSYRGHTDEVCGLKWSLDGKKLASGGSDNVVHIW 742
Query: 124 DTRERSCCNSFEAHEDYISDITFASDAMKLLA 155
D RS +S ++ + A+K LA
Sbjct: 743 D---RSAVSSSSRTTRWLHKFEEHTAAVKALA 829
>TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {Arabidopsis
thaliana}, partial (93%)
Length = 2633
Score = 40.0 bits (92), Expect = 0.001
Identities = 18/68 (26%), Positives = 37/68 (53%)
Frame = +3
Query: 95 AHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLL 154
AH+ + ++ L + +G D +K W + ++C ++F+ H D + + SD + +L
Sbjct: 525 AHKGPIQAVIKLPTGELVTGSSDTTLKTW--KGKTCLHTFQGHADTVRGLAVMSD-LGIL 695
Query: 155 ATSGDGTL 162
+ S DG+L
Sbjct: 696 SASHDGSL 719
>TC78038 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Arabidopsis
thaliana}, partial (60%)
Length = 1986
Score = 40.0 bits (92), Expect = 0.001
Identities = 49/231 (21%), Positives = 96/231 (41%), Gaps = 22/231 (9%)
Frame = +3
Query: 64 NGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLV-NLTESTVASGDDDGCIKV 122
+ G L+ G I +VETG + + + H AV LV + +S + SG +G ++V
Sbjct: 441 HSGTYLVGGGLSGEIYLWEVETGRLLKKW-HGHNTAVTCLVFSEDDSLLISGFKNGSVRV 617
Query: 123 W-------DTRERSCCN----SFEAHEDYISDITFASDAMK--LLATSGDGTLSVCNLRR 169
W D R R SF H ++D+ + + S D T V L
Sbjct: 618 WSLLMIFDDVRRREASKIYEYSFSDHTLCVNDVVIGYGGCNAIIASASDDRTCKVWTLSN 797
Query: 170 NKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDT 229
+Q F ++ ++ L + GS+ G + + + + ++ V + S
Sbjct: 798 GMLQRNIVF-PSKIKAIALDPAEHVLYAGSEDGKIFVAALNTTRITTNDQVSYITGSFSN 974
Query: 230 MLK--------LDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERL 272
K E+ +I+GS++G++ + + +I+ V +H++ P+ +
Sbjct: 975 HSKAVTCLAYSAAENFLISGSDDGIVRVWNASTHNIIR-VFKHAKGPITNI 1124
>BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [imported] -
Arabidopsis thaliana, partial (15%)
Length = 654
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/86 (27%), Positives = 42/86 (47%), Gaps = 1/86 (1%)
Frame = +1
Query: 82 DVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDY 140
D G+ I + D H+ V + + ++ SG DD IKVW+ + C + H DY
Sbjct: 181 DYRMGTLIDKFDE-HDGPVRGVHFHNSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDY 357
Query: 141 ISDITFASDAMKLLATSGDGTLSVCN 166
I + F ++ +++ S D T+ + N
Sbjct: 358 IRTVQFHHESPWIVSASDDQTIRIWN 435
Score = 38.5 bits (88), Expect = 0.004
Identities = 32/173 (18%), Positives = 65/173 (37%)
Frame = +1
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ FHP + L G + L+ Y + + H R F N ++
Sbjct: 115 LSFHPKRPWILASLHSGVIQLWDYRMGTL------IDKFDEHDGPVRGVHFHNSQPLFVS 276
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCC 131
G D+ I + + + L + + + S DD I++W+ + R+C
Sbjct: 277 GGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCI 456
Query: 132 NSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL 184
+ H Y+ F +++ S D T+ V ++ ++ +S D++L
Sbjct: 457 SVLTGHNHYVMCALFHPKDDLVVSASLDQTVRVWDI--GSLKRKSASXADDIL 609
>TC90247 weakly similar to GP|21592898|gb|AAM64848.1 unknown {Arabidopsis
thaliana}, partial (60%)
Length = 1041
Score = 38.9 bits (89), Expect = 0.003
Identities = 53/230 (23%), Positives = 99/230 (43%), Gaps = 23/230 (10%)
Frame = +1
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLV-NLTESTVASGDDDGCIKVW- 123
G L+ G+ I +VETG + + +AH AV LV + +S + SG +DG ++VW
Sbjct: 352 GTYLVGGALSGDIYLWEVETGRLLKKW-HAHLRAVTCLVFSDDDSLLISGSEDGYVRVWS 528
Query: 124 ------DTRERSCCN----SFEAHEDYISDITFASDAMK--LLATSGDGTLSVCNLRRNK 171
D R R N SF H ++D+ + +++ S D T V +L R
Sbjct: 529 LFSLFDDLRSREEKNLYEYSFSEHTMRVTDVVIGNGGCNAIIVSASDDRTCKVWSLSRGT 708
Query: 172 VQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWG-----CFKDCSDRFVDLSSN- 225
+ F + ++VL GS G + + + + + SN
Sbjct: 709 LLRSIVF-PSVINAIVLDPAEHVFYAGSSDGKIFIAALNTERITTIDNYGMHIIGSFSNH 885
Query: 226 -SIDTMLKLDEDR--IITGSENGMINLVGILPNRVIQPVAEHSEYPVERL 272
T L + R +I+GSE+G++ + + +++ + +H++ P+ +
Sbjct: 886 SKAVTCLAYSKGRNLLISGSEDGIVRVWNAKTHNIVR-MFKHAKGPITNI 1032
>TC80645 similar to PIR|T50983|T50983 probable pleiotropic regulator 1
(PLRG1) [imported] - Neurospora crassa, partial (41%)
Length = 1141
Score = 38.9 bits (89), Expect = 0.003
Identities = 32/139 (23%), Positives = 61/139 (43%), Gaps = 9/139 (6%)
Frame = +2
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRER 128
++TGS D ++ D+ G ++ L + H+ V L E + G IK W E
Sbjct: 125 VITGSLDATVRLWDLAAGKSMGVLTH-HKKGVRALATHPEEFTFASGSSGSIKQWKCPEG 301
Query: 129 SCCNSFEAHEDYISDITFASDAMKLLATSGD-GTLSVCNLR--------RNKVQAQSEFS 179
+ +FE H I+ ++ D +L + GD G+++ + + Q S +
Sbjct: 302 AFMQNFEGHNAIINTLSVNRD--NVLFSGGDNGSMNFWDWKTGHRFQSLETTAQPGSLDA 475
Query: 180 EDELLSVVLMKNGRKVVCG 198
E ++S +G +++CG
Sbjct: 476 EAGIMSSTFDNSGLRLICG 532
Score = 34.3 bits (77), Expect = 0.081
Identities = 29/134 (21%), Positives = 55/134 (40%), Gaps = 1/134 (0%)
Frame = +2
Query: 116 DDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQ 175
+ G +VWD R RS + H +SD+T +++ S D T+ + +L K
Sbjct: 14 ETGLPRVWDMRTRSNVHVLSGHTGTVSDLTCQEADPQVITGSLDATVRLWDLAAGKSMGV 193
Query: 176 SEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDE 235
+ + ++ GS I W C + + + N+I L ++
Sbjct: 194 LTHHKKGVRALATHPEEFTFASGSSGSIK---QWKCPEGAFMQNFE-GHNAIINTLSVNR 361
Query: 236 DRII-TGSENGMIN 248
D ++ +G +NG +N
Sbjct: 362 DNVLFSGGDNGSMN 403
>TC90611 similar to GP|10177223|dbj|BAB10298. contains similarity to
GTP-binding regulatory protein and WD-repeat
protein~gene_id:MPF21.14, partial (40%)
Length = 855
Score = 38.5 bits (88), Expect = 0.004
Identities = 29/122 (23%), Positives = 53/122 (42%), Gaps = 10/122 (8%)
Frame = +1
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVA 112
H ++ + G+ L + S D +I + + + + NAH+ A+N +V + T
Sbjct: 478 HVDTVSSLALSKDGKFLYSVSWDRTIKIWRTKDFTCLESITNAHDDAINAVVVSYDGTFY 657
Query: 113 SGDDDGCIKVWDT----------RERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTL 162
SG D IKVW + S ++ E H I+ + +SD L + + D ++
Sbjct: 658 SGSADKRIKVWKNLQGDDHNKKKHKHSLVDTLEKHNSGINALVLSSDESILYSGACDRSI 837
Query: 163 SV 164
V
Sbjct: 838 LV 843
>CA917664 similar to GP|16604679|gb putative WD-repeat membrane protein
{Arabidopsis thaliana}, partial (24%)
Length = 749
Score = 37.7 bits (86), Expect = 0.007
Identities = 19/54 (35%), Positives = 31/54 (57%)
Frame = +2
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
+A+ DD I+++D FE H D I+D+ F+ D LL++S DG+L +
Sbjct: 164 LATVADDLTIRLFDVVALRLVRKFEGHTDRITDLCFSEDGKWLLSSSMDGSLRI 325
>TC78328 similar to GP|11994333|dbj|BAB02292. WD-40 repeat protein-like
{Arabidopsis thaliana}, partial (88%)
Length = 1516
Score = 37.4 bits (85), Expect = 0.010
Identities = 27/104 (25%), Positives = 43/104 (40%)
Frame = +1
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDT 125
G L++ S D S + + ETG I + A + ++ A+ D KVWD
Sbjct: 352 GFFLISASKDASPMLRNGETGDWIGTFEGHKGAVWSCCLDANALRAATASADFSTKVWDA 531
Query: 126 RERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRR 169
+SFE H+ + F+ D LL + L + +L R
Sbjct: 532 LTGDALHSFE-HKHIVRACAFSEDTHLLLTGGAEKILRIYDLNR 660
Score = 30.0 bits (66), Expect = 1.5
Identities = 18/61 (29%), Positives = 28/61 (45%), Gaps = 1/61 (1%)
Frame = +1
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWD 124
G + G D + D TG IA + H V+ + + + ASG +DG I++W
Sbjct: 979 GNKFVAGGEDLWVHVFDFHTGDEIA-CNKGHHGPVHCVRFSPGGESYASGSEDGTIRIWQ 1155
Query: 125 T 125
T
Sbjct: 1156 T 1158
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,006,119
Number of Sequences: 36976
Number of extensions: 121525
Number of successful extensions: 840
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 754
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 819
length of query: 378
length of database: 9,014,727
effective HSP length: 98
effective length of query: 280
effective length of database: 5,391,079
effective search space: 1509502120
effective search space used: 1509502120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC142222.5


