

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142142.4 - phase: 0
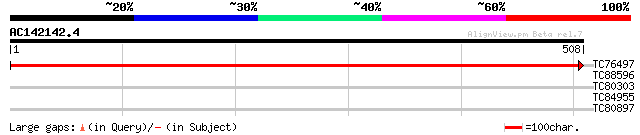
(508 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76497 homologue to SP|P06411|PSBB_TOBAC Photosystem II P680 ch... 1060 0.0
TC88596 homologue to GP|4581045|gb|AAD24583.1| chlorophyll a-bin... 41 0.001
TC80303 weakly similar to GP|20161135|dbj|BAB90063. hypothetical... 28 6.4
TC84955 similar to GP|9716226|emb|CAC01587.1 putative lipoprotei... 28 8.3
TC80897 similar to GP|3241854|dbj|BAA28872.1 thaumatin-like prot... 28 8.3
>TC76497 homologue to SP|P06411|PSBB_TOBAC Photosystem II P680 chlorophyll A
apoprotein (CP-47 protein). [Common tobacco] {Nicotiana
tabacum}, complete
Length = 4045
Score = 1060 bits (2742), Expect = 0.0
Identities = 508/508 (100%), Positives = 508/508 (100%)
Frame = +1
Query: 1 MGLPWYRVHTVVLNDPGRLLSVHIMHTALVAGWAGSMALYELAVFDPSDPVLDPMWRQGM 60
MGLPWYRVHTVVLNDPGRLLSVHIMHTALVAGWAGSMALYELAVFDPSDPVLDPMWRQGM
Sbjct: 145 MGLPWYRVHTVVLNDPGRLLSVHIMHTALVAGWAGSMALYELAVFDPSDPVLDPMWRQGM 324
Query: 61 FVIPFMTRLGITNSWGGWNITGGTITNPGIWSYEGVAAAHIVFSGLCFLAAIWHWVYWDL 120
FVIPFMTRLGITNSWGGWNITGGTITNPGIWSYEGVAAAHIVFSGLCFLAAIWHWVYWDL
Sbjct: 325 FVIPFMTRLGITNSWGGWNITGGTITNPGIWSYEGVAAAHIVFSGLCFLAAIWHWVYWDL 504
Query: 121 EIFCDERTGKPSLDLPKIFGIHLFLAGVACFGFGAFHVTGLFGPGIWVSDPYGLTGRVQS 180
EIFCDERTGKPSLDLPKIFGIHLFLAGVACFGFGAFHVTGLFGPGIWVSDPYGLTGRVQS
Sbjct: 505 EIFCDERTGKPSLDLPKIFGIHLFLAGVACFGFGAFHVTGLFGPGIWVSDPYGLTGRVQS 684
Query: 181 VNPAWGVDGFDPFVPGGIASHHIAAGTLGILAGLFHLSVRPPQRLYKGLRMGNIETVLSS 240
VNPAWGVDGFDPFVPGGIASHHIAAGTLGILAGLFHLSVRPPQRLYKGLRMGNIETVLSS
Sbjct: 685 VNPAWGVDGFDPFVPGGIASHHIAAGTLGILAGLFHLSVRPPQRLYKGLRMGNIETVLSS 864
Query: 241 SIAAVFFAAFVVAGTMWYGSATTPIELFGPTRYQWDQGYFQQEIYRRVGAGLAENQSLSE 300
SIAAVFFAAFVVAGTMWYGSATTPIELFGPTRYQWDQGYFQQEIYRRVGAGLAENQSLSE
Sbjct: 865 SIAAVFFAAFVVAGTMWYGSATTPIELFGPTRYQWDQGYFQQEIYRRVGAGLAENQSLSE 1044
Query: 301 AWSKIPEKLAFYDYIGNNPAKGGLFRAGSMDNGDGIAVGWLGHPIFRDKEGRELFVRRMP 360
AWSKIPEKLAFYDYIGNNPAKGGLFRAGSMDNGDGIAVGWLGHPIFRDKEGRELFVRRMP
Sbjct: 1045AWSKIPEKLAFYDYIGNNPAKGGLFRAGSMDNGDGIAVGWLGHPIFRDKEGRELFVRRMP 1224
Query: 361 TFFETFPVVLVDGDGIVRADVPFRRAESKYSVEQVGVTVEFFGGELNGVSYSDPATVKKY 420
TFFETFPVVLVDGDGIVRADVPFRRAESKYSVEQVGVTVEFFGGELNGVSYSDPATVKKY
Sbjct: 1225TFFETFPVVLVDGDGIVRADVPFRRAESKYSVEQVGVTVEFFGGELNGVSYSDPATVKKY 1404
Query: 421 ARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGHASFALLFFFGHIWHGARTLFRDVFA 480
ARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGHASFALLFFFGHIWHGARTLFRDVFA
Sbjct: 1405ARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGHASFALLFFFGHIWHGARTLFRDVFA 1584
Query: 481 GIDPDLDAQVEFGAFQKLGDPTTKKQAV 508
GIDPDLDAQVEFGAFQKLGDPTTKKQAV
Sbjct: 1585GIDPDLDAQVEFGAFQKLGDPTTKKQAV 1668
>TC88596 homologue to GP|4581045|gb|AAD24583.1| chlorophyll a-binding
protein PsbC {Populus deltoides}, partial (37%)
Length = 1650
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/73 (28%), Positives = 29/73 (38%)
Frame = +3
Query: 419 KYARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGHASFALLFFFGHIWHGARTLFRDV 478
+Y A LG + + + + V SPR W H F GH+WH R R
Sbjct: 294 EYMTHAPLGSLNSVGGVATEINAVNYVSPRSWLATSHFVLGFFLFVGHLWHAGRA--RAA 467
Query: 479 FAGIDPDLDAQVE 491
AG + +D E
Sbjct: 468 AAGFEKGIDRDSE 506
>TC80303 weakly similar to GP|20161135|dbj|BAB90063. hypothetical protein
{Oryza sativa (japonica cultivar-group)}, partial (19%)
Length = 789
Score = 28.5 bits (62), Expect = 6.4
Identities = 13/25 (52%), Positives = 14/25 (56%)
Frame = +1
Query: 143 LFLAGVACFGFGAFHVTGLFGPGIW 167
LFL V CF F TG+FG G W
Sbjct: 70 LFLLTVVCFILVLFRRTGMFGRGEW 144
>TC84955 similar to GP|9716226|emb|CAC01587.1 putative lipoprotein
{Streptomyces coelicolor A3(2)}, partial (2%)
Length = 650
Score = 28.1 bits (61), Expect = 8.3
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = -2
Query: 133 LDLPKIFGIHLFLAGVACFGFGAFHVTGLFGPGIWVS 169
LDL + G +GV G FH+ FGPG++ S
Sbjct: 145 LDLA*LKGGAPLNSGVTLTSIGKFHLPSAFGPGLYTS 35
>TC80897 similar to GP|3241854|dbj|BAA28872.1 thaumatin-like protein
precursor {Pyrus pyrifolia}, partial (60%)
Length = 602
Score = 28.1 bits (61), Expect = 8.3
Identities = 15/49 (30%), Positives = 20/49 (40%), Gaps = 7/49 (14%)
Frame = -3
Query: 78 WNITGGTITNPGIWSY-----EGVAAAHIV--FSGLCFLAAIWHWVYWD 119
W G +T P +W Y E AHIV S +C + +WD
Sbjct: 189 WTTGGFAVTTPTLWRYRHWHVESTHIAHIVKILSPVCCYGYFYQCCWWD 43
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.143 0.469
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,699,083
Number of Sequences: 36976
Number of extensions: 254446
Number of successful extensions: 1343
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 1334
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1343
length of query: 508
length of database: 9,014,727
effective HSP length: 100
effective length of query: 408
effective length of database: 5,317,127
effective search space: 2169387816
effective search space used: 2169387816
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142142.4


