

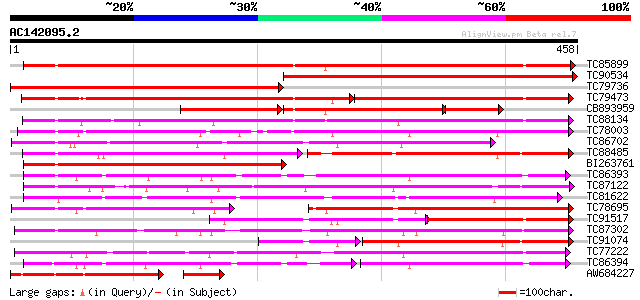
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142095.2 + phase: 0
(458 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:... 491 e-139
TC90534 weakly similar to GP|19911197|dbj|BAB86925. glucosyltran... 491 e-139
TC79736 weakly similar to PIR|B84871|B84871 probable glucosyltra... 456 e-129
TC79473 weakly similar to PIR|T00639|T00639 hypothetical protein... 200 e-100
CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic ... 176 5e-80
TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 282 2e-76
TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase... 272 2e-73
TC86702 weakly similar to GP|9794913|gb|AAF98390.1| UDP-glucose:... 226 1e-59
TC88485 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 167 2e-55
BI263761 similar to GP|7385017|gb|A UDP-glucose:salicylic acid g... 205 3e-53
TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyl... 201 5e-52
TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltran... 189 2e-48
TC81622 weakly similar to PIR|T00584|T00584 indole-3-acetate bet... 189 2e-48
TC78695 similar to GP|4115563|dbj|BAA36423.1 UDP-glucose:anthocy... 188 3e-48
TC91517 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 113 6e-45
TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase... 177 6e-45
TC91074 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 152 2e-44
TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 174 8e-44
TC86394 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyl... 109 9e-42
AW684227 similar to GP|7385017|gb|A UDP-glucose:salicylic acid g... 150 3e-41
>TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:salicylic
acid glucosyltransferase {Nicotiana tabacum}, partial
(47%)
Length = 1617
Score = 491 bits (1265), Expect = e-139
Identities = 234/448 (52%), Positives = 325/448 (72%), Gaps = 2/448 (0%)
Frame = +2
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDGF 71
HCL+LP+PA GH NPM++FSK L ++ GVK+TL+T S +K I SI +E+ISDG+
Sbjct: 44 HCLILPYPAQGHMNPMIQFSKRLIEK-GVKITLITVTSFWKVISNKNLTSIDVESISDGY 220
Query: 72 DKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEFG 131
D+GG+ A+ + Y FW+VG Q+L+ L++ L++ + +C+I+D+F+PW LDV K FG
Sbjct: 221 DEGGLLAAESLEDYKETFWKVGSQTLSELLHKLSSSENPPNCVIFDAFLPWVLDVGKSFG 400
Query: 132 IVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQDPTF 191
+VG +F TQ+ +NS+YYH H ++ P + E LP LP+L P D+PSF + Y P +
Sbjct: 401 LVGVAFFTQSCSVNSVYYHTHEKLIELPLTQSEYLLPGLPKLAPGDLPSFLYKYGSYPGY 580
Query: 192 LDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRVKDDE 251
DI V QF+NI KADWIL NS +ELE EV DW +KIW +T+GP +P LDKR+KDD+
Sbjct: 581 FDIVVNQFANIGKADWILANSIYELEPEVVDWLVKIWP-LKTIGPSVPSMLLDKRLKDDK 757
Query: 252 DH--SIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVV 309
++ S++ ++ I+WLN+KPK S VY SFGSMA L+EEQ +E+A LKD SYFLWVV
Sbjct: 758 EYGVSLSDPNTEFCIKWLNDKPKGSVVYASFGSMAGLSEEQTQELALGLKDSESYFLWVV 937
Query: 310 KTSEETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVA 369
+ +++KLPK F + S+ GL+V WCPQL VL HEA+GCFVTHCGWNSTLEALSIGVP++A
Sbjct: 938 RECDQSKLPKGFVESSKKGLIVTWCPQLLVLTHEALGCFVTHCGWNSTLEALSIGVPLIA 1117
Query: 370 IPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQ 429
+PL++DQ +AK + D+WK+G+R + DEK+IVR + +K+CI EI+ +EKG EI N ++
Sbjct: 1118MPLWTDQVTNAKLIADVWKMGVRAVADEKEIVRSETIKNCIKEIIE-TEKGNEIKKNALK 1294
Query: 430 WKTLATRAVGKDGSSHKNMIEFVNSLFQ 457
WK LA +V + G S KN+ EFV +L Q
Sbjct: 1295WKNLAKSSVDEGGRSDKNIEEFVAALAQ 1378
>TC90534 weakly similar to GP|19911197|dbj|BAB86925. glucosyltransferase-7
{Vigna angularis}, partial (71%)
Length = 808
Score = 491 bits (1264), Expect = e-139
Identities = 236/237 (99%), Positives = 237/237 (99%)
Frame = +3
Query: 222 DWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFG 281
DWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFG
Sbjct: 3 DWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFG 182
Query: 282 SMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLVVAWCPQLEVLA 341
SMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGL+VAWCPQLEVLA
Sbjct: 183 SMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLIVAWCPQLEVLA 362
Query: 342 HEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIV 401
HEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIV
Sbjct: 363 HEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIV 542
Query: 402 RKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSLFQV 458
RKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSLFQV
Sbjct: 543 RKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSLFQV 713
>TC79736 weakly similar to PIR|B84871|B84871 probable glucosyltransferase
[imported] - Arabidopsis thaliana, partial (8%)
Length = 1055
Score = 456 bits (1173), Expect = e-129
Identities = 219/221 (99%), Positives = 221/221 (99%)
Frame = +1
Query: 1 MENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNN 60
MENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNN
Sbjct: 46 MENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNN 225
Query: 61 SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFM 120
SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFM
Sbjct: 226 SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFM 405
Query: 121 PWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPS 180
PWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQP+DMPS
Sbjct: 406 PWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPKDMPS 585
Query: 181 FYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVA 221
FYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEV+
Sbjct: 586 FYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVS 708
>TC79473 weakly similar to PIR|T00639|T00639 hypothetical protein F3I6.2 -
Arabidopsis thaliana, partial (45%)
Length = 1530
Score = 200 bits (509), Expect(2) = e-100
Identities = 107/272 (39%), Positives = 170/272 (62%), Gaps = 3/272 (1%)
Frame = +2
Query: 10 SVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISD 69
+VH LV+P+PA GH +P+++FSK L + G+K T TT K+I PN I++E ISD
Sbjct: 68 NVHVLVIPYPAQGHISPLIQFSKRLVSK-GIKTTFATTHYTVKSITA-PN--ISVEPISD 235
Query: 70 GFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKE 129
GFD+ G ++AK+ +L+LN F G ++L++LI + + C++YDSF+PW LDVAK+
Sbjct: 236 GFDESGFSQAKNVELFLNSFKTNGSKTLSNLIQKHQKTSTPITCIVYDSFLPWALDVAKQ 415
Query: 130 FGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQDP 189
I GA+F T + + +I+ +H G ++ P E + +P LP L RD+PSF E P
Sbjct: 416 HRIYGAAFFTNSAAVCNIFCRIHHGLIETPVDELPLIVPGLPPLNSRDLPSFIRFPESYP 595
Query: 190 TFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRVKD 249
++ + + QFSN+++ADW+ N+F LE EV +++ + +GP +P +LD R+K
Sbjct: 596 AYMAMKLNQFSNLNQADWMFVNTFEALEAEVVKGLTEMFP-AKLIGPMVPSAYLDGRIKG 772
Query: 250 DEDHSIAQLK--SDESIEWLNNKPKRS-AVYV 278
D+ + K S++ I WLN KP +S +VY+
Sbjct: 773 DKGYGANLWKPLSEDCINWLNAKPSQSVSVYL 868
Score = 183 bits (465), Expect(2) = e-100
Identities = 92/178 (51%), Positives = 126/178 (70%), Gaps = 1/178 (0%)
Frame = +1
Query: 279 SFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKK-SENGLVVAWCPQL 337
SFGSM SL EQIEE+A LK+ FLWV++ SE+ KLPK ++ E G++V WC QL
Sbjct: 868 SFGSMVSLTSEQIEELALGLKESEVNFLWVLRESEQGKLPKGYKDFIKEKGIIVTWCNQL 1047
Query: 338 EVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDE 397
E+LAH+A+GCFVTHCGWNSTLE+LS+GVP+V +P ++DQ DAKFL +IW+VG+RP DE
Sbjct: 1048 ELLAHDAVGCFVTHCGWNSTLESLSLGVPVVCLPQWADQLPDAKFLEEIWEVGVRPKEDE 1227
Query: 398 KQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+V+++ + +M SE+ + I N +WK LA AV + GS KN+ +FV+ L
Sbjct: 1228 NGVVKREEFMLSLKVVME-SERSEVIRRNASEWKKLARDAVCEGGSFDKNINQFVDYL 1398
>CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic acid
glucosyltransferase {Nicotiana tabacum}, partial (28%)
Length = 791
Score = 176 bits (445), Expect(3) = 5e-80
Identities = 82/134 (61%), Positives = 103/134 (76%), Gaps = 2/134 (1%)
Frame = +3
Query: 222 DWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDH--SIAQLKSDESIEWLNNKPKRSAVYVS 279
DW KIW +T+GP +P + LDKR+KDD+++ S++ ++ SI+WLN KPKRS VYVS
Sbjct: 252 DWLSKIWP-LKTIGPSVPSSHLDKRIKDDKEYGVSVSDPNTESSIKWLNEKPKRSVVYVS 428
Query: 280 FGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLVVAWCPQLEV 339
FGS A L+EEQIEE+A L D YFLWVV+ SE+ KLPK FE+ S+NGL+V WCPQLEV
Sbjct: 429 FGSNARLSEEQIEELALGLNDSEKYFLWVVRESEQVKLPKGFEETSKNGLIVTWCPQLEV 608
Query: 340 LAHEAIGCFVTHCG 353
L HEA+ CFVTHCG
Sbjct: 609 LTHEAVACFVTHCG 650
Score = 86.7 bits (213), Expect(3) = 5e-80
Identities = 41/82 (50%), Positives = 53/82 (64%)
Frame = +2
Query: 139 TQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQ 198
TQ+ +NSIY+H H ++ P + E LP LP+L D+PSF + Y P D+ V Q
Sbjct: 2 TQSCSVNSIYFHTHQKLIELPISQSEYLLPGLPKLAQGDLPSFLYKYGSYPIIFDVVVNQ 181
Query: 199 FSNIHKADWILCNSFFELEKEV 220
FSNI KADWIL N+F+ELE EV
Sbjct: 182 FSNIGKADWILANTFYELEPEV 247
Score = 75.5 bits (184), Expect(3) = 5e-80
Identities = 30/47 (63%), Positives = 42/47 (88%)
Frame = +1
Query: 353 GWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQ 399
GWNSTLEALSIGVP++A+PL++DQ +AKF+ D+WK+G+R + DEK+
Sbjct: 649 GWNSTLEALSIGVPLIAMPLWTDQATNAKFIADVWKMGVRAVADEKE 789
>TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (27%)
Length = 1669
Score = 282 bits (721), Expect = 2e-76
Identities = 171/467 (36%), Positives = 254/467 (53%), Gaps = 22/467 (4%)
Frame = +2
Query: 11 VHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPK------------LP 58
+H L++ F A GH NP+L K L + G+ VTL TT Y + K
Sbjct: 71 IHVLLVAFSAQGHINPLLRLGKSLLTK-GLNVTLATTELVYHRVFKSTTTDTDTVPTSYS 247
Query: 59 NNSITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINN--LNARNDHVDCLIY 116
N I + SDGFD ++ D Y+ + GP SL +LINN +N + + C+I
Sbjct: 248 TNGINVLFFSDGFDISQGHKSPDE--YMELIAKFGPISLTNLINNNFINNPSKKLACIIN 421
Query: 117 DSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQ---EITLPALPQL 173
+ F+PW +VA E I A Q + SIYY + + P +E ++ LP LP L
Sbjct: 422 NPFVPWVANVAYELNIPCACLWIQPCTLYSIYYRFYNELNQFPTLENPEIDVELPGLPLL 601
Query: 174 QPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRT 233
+P+D+PSF T D+ F ++ K W+L NSF+ELEK+V D +I+
Sbjct: 602 KPQDLPSFILPSNPIKTMSDVLAEMFQDMKKLKWVLANSFYELEKDVIDSMAEIFP-ITP 778
Query: 234 VGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEE 293
VGP +P + L + K D + D +EWLN K S +Y+SFGS+ L+E+Q++
Sbjct: 779 VGPLVPPSLLGQDQKQDVGIEM-WTPQDSCMEWLNQKLPSSVIYISFGSLIFLSEKQMQS 955
Query: 294 VAHCLKDCGSYFLWVVKTS----EETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCF 348
+A LK+ YFLWV+K++ E+ +L + F E+ E GLVV WCPQ +VL H AI CF
Sbjct: 956 IASALKNSNKYFLWVMKSNDIGAEKVQLSEKFLEETKEKGLVVTWCPQTKVLVHPAIACF 1135
Query: 349 VTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKD 408
+THCGWNSTLEA++ GVP++ P ++DQ +A + D++++GIR D V D ++
Sbjct: 1136LTHCGWNSTLEAITAGVPMIGYPQWTDQPTNATLVSDVFRMGIRLKQDNDGFVESDEVER 1315
Query: 409 CICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
I EI+ E+ + N ++ K A AV GSS +N+ FV+ +
Sbjct: 1316AIEEIVG-GERSEVFKKNALELKHAAREAVADGGSSDRNIQRFVDEI 1453
>TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase NTGT2
{Nicotiana tabacum}, partial (50%)
Length = 1740
Score = 272 bits (696), Expect = 2e-73
Identities = 174/478 (36%), Positives = 257/478 (53%), Gaps = 29/478 (6%)
Frame = +3
Query: 7 STKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNI---PKLPNNSIT 63
ST L++P+P GH NP EF+K L G VT+ TT+ + I P LPN ++
Sbjct: 135 STMPHRILLVPYPVQGHINPAFEFAKRLITL-GAHVTISTTVHMHNRITNKPTLPN--LS 305
Query: 64 IETISDGFDKGGVAEAKDFKL-YLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPW 122
SDG+D G D L Y +F + G + ++ +I + CL++ + W
Sbjct: 306 YYPFSDGYDDGFKGTGSDAYLEYHAEFQRRGSEFVSDIILKNSQEGTPFTCLVHSLLLQW 485
Query: 123 CLDVAKEFGIVGASFLTQNL-VMNSIYYHVH--LGKLKPPFVEQEITLPALPQL-QPRDM 178
+ A+EF + A Q V + +YY+ H +K P I LP LP L RD+
Sbjct: 486 AAEAAREFHLPTALLWVQPATVFDILYYYFHGFSDSIKNP--SSSIELPGLPLLFSSRDL 659
Query: 179 PSFYFT------------YEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMK 226
PSF +E+ LD+ +N+ K IL NSF LE + K
Sbjct: 660 PSFLLASCPDAYSLMTSFFEEQFNELDVE----TNLTKT--ILVNSFESLEPKALRAVKK 821
Query: 227 IWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLK----SDESIEWLNNKPKRSAVYVSFGS 282
N ++GP +P LD++ +++ Q S++ +EWL++KPK S VYVSFGS
Sbjct: 822 F--NMISIGPLIPSEHLDEKDSTEDNSYGGQTHIFQPSNDCVEWLDSKPKSSVVYVSFGS 995
Query: 283 MASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDF---EKKSENGLVVAWCPQLEV 339
L+E Q EE+AH L DCG FLWV++ E + F E+ E G +V WC Q+E+
Sbjct: 996 YFVLSERQREEIAHALLDCGFPFLWVLREKEGENNEEGFKYREELEEKGKIVKWCSQMEI 1175
Query: 340 LAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR--PLVDE 397
L+H ++GCF+THCGWNSTLE+L GVP+VA P ++DQ +AK + D+WK+G+R V+E
Sbjct: 1176LSHPSLGCFLTHCGWNSTLESLVKGVPMVAFPQWTDQMTNAKLIEDVWKIGVRVDEEVNE 1355
Query: 398 KQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
IVR D ++ C+ +M EKG+E+ + +WK LA AV + GSS KN+ F++ +
Sbjct: 1356DGIVRGDEIRRCLEVVMGSGEKGEELRRSGKKWKELAREAVKEGGSSEKNLRSFLDGV 1529
>TC86702 weakly similar to GP|9794913|gb|AAF98390.1| UDP-glucose:sinapate
glucosyltransferase {Brassica napus}, partial (34%)
Length = 1366
Score = 226 bits (576), Expect = 1e-59
Identities = 143/416 (34%), Positives = 216/416 (51%), Gaps = 25/416 (6%)
Frame = +3
Query: 2 ENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTT---------ISNY- 51
+ K S + H L++ +PA GH NP+L +K L + G V +TT ++N
Sbjct: 48 KKKMGSEANTHILLISYPAQGHINPLLRLAKCLAAK-GASVIFITTEKAGKDIRTVNNII 224
Query: 52 -KNIPKLPNNSITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDH 110
K++ + + S+T E + DG + + Y+ VG ++ +I N N
Sbjct: 225 EKSLTSIGDGSLTFEFLDDGLEDDHPLRG-NLLGYIEHLKLVGKPFVSQMIKNHADSNKP 401
Query: 111 VDCLIYDSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYH-----VHLGKLKPPFVEQEI 165
C+I + F+PW DVA E GI A Q+ + S YY+ V P+++ ++
Sbjct: 402 FSCIINNPFVPWVCDVADEHGIPSALLWIQSTAVFSAYYNYFHKLVRFPSQTEPYIDVQL 581
Query: 166 TLPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTM 225
L ++P F + Q P + + QF N+ K +L ++ ELE + D+
Sbjct: 582 PFQVLKY---NEIPDFLHPFSQFPFLGTLILEQFKNLSKVFCVLVETYEELEHDFIDYIS 752
Query: 226 KIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDES--IEWLNNKPKRSAVYVSFGSM 283
K R +GP F + +K ++ +KSD+ IEWL+ KPK S VY+SFGS+
Sbjct: 753 KKTIFVRPIGPL----FHNPNIKGAKNIRGDFVKSDDCNIIEWLSTKPKGSVVYISFGSI 920
Query: 284 ASLNEEQIEEVAHCLKDCGSYFLWVVKTSEET------KLPKDF-EKKSENGLVVAWCPQ 336
+ +EQ+ E+A+ L D FLWV+K E LP F E SE G VV W PQ
Sbjct: 921 VYIPQEQVNEIAYGLLDSQVSFLWVLKPPSEELGLKEHNLPDGFLEGISERGKVVNWSPQ 1100
Query: 337 LEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR 392
EVLAH ++ CF+THCGWNS++EALS+GVP++ P + DQ +AKFLVD++ VGIR
Sbjct: 1101EEVLAHPSVACFITHCGWNSSMEALSLGVPMLTFPAWGDQVTNAKFLVDVFGVGIR 1268
>TC88485 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (39%)
Length = 1544
Score = 167 bits (422), Expect(2) = 2e-55
Identities = 89/219 (40%), Positives = 135/219 (61%), Gaps = 4/219 (1%)
Frame = +2
Query: 241 TFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKD 300
TF+ K ++D D+ IEWL+ KPK S VYVSFG++ + +EQ+ E+ + L +
Sbjct: 812 TFMVDFAKSNDD--------DKIIEWLDTKPKDSVVYVSFGTLVNYPQEQMNEIVYGLLN 967
Query: 301 CGSYFLWVVKTSEETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLE 359
FLW + S LP DF E+ +E G VV W PQ++VLAH ++ CF+THCGWNS++E
Sbjct: 968 SQVSFLWSL--SNPGVLPDDFLEETNERGKVVEWSPQVDVLAHPSVACFITHCGWNSSIE 1141
Query: 360 ALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR---PLVDEKQIVRKDPLKDCICEIMSM 416
ALS+GVP++ P DQ +AKFLVD++ VGI+ ++V +D +K C+ E ++
Sbjct: 1142 ALSLGVPVLTFPSRGDQLTNAKFLVDVFGVGIKMGSGWAVNNKLVTRDEVKKCLLE-ATI 1318
Query: 417 SEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
EK +++ N +WK A A+ GSS +N+ EF+ +
Sbjct: 1319 GEKAEKLKQNANKWKKKAEEALAVGGSSDRNLDEFMEDI 1435
Score = 67.4 bits (163), Expect(2) = 2e-55
Identities = 65/239 (27%), Positives = 101/239 (42%), Gaps = 13/239 (5%)
Frame = +1
Query: 11 VHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIET-ISD 69
+ L++ FPA GH N ++ K L + G V TT + KN+ N + T I D
Sbjct: 64 IKLLLVSFPAQGHINHLVGLGKYLAAK-GATVIFTTTETAGKNMRAANNIIDKLATPIGD 240
Query: 70 G------FDKG---GVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFM 120
G FD G G A + + G S++ +I N N C+I + F
Sbjct: 241 GTFAFEFFDDGLPDGDRSAFRALQHSAEIEVAGRPSISQMIKNHADLNKPFSCIINNYFF 420
Query: 121 PWCLDVAKEFGIVGASFLTQNLVMNSIYY-HVHLGKLKPPFVEQEITLPALPQ--LQPRD 177
PW DVA E I T + + + YY +VH P E I + +P L+ +
Sbjct: 421 PWVCDVANEHNIPSVLSWTNSAAVFTTYYNYVHKLTPFPTNEEPYIDVQLIPSRVLKYNE 600
Query: 178 MPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGP 236
+ + P + + +F ++ K +L +++ ELE E D+ K RTVGP
Sbjct: 601 ISDLVHPFCSFPFLGKLVLEEFKDLSKVFCVLVDTYEELEHEFIDYISKKSIPIRTVGP 777
>BI263761 similar to GP|7385017|gb|A UDP-glucose:salicylic acid
glucosyltransferase {Nicotiana tabacum}, partial (6%)
Length = 666
Score = 205 bits (521), Expect = 3e-53
Identities = 101/213 (47%), Positives = 141/213 (65%), Gaps = 1/213 (0%)
Frame = +2
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPK-LPNNSITIETISDG 70
H LV P+P GH NPML+FSK L Q+ GVK+TLV TIS + I + NSI E+ISDG
Sbjct: 23 HILVFPYPLQGHINPMLQFSKRLSQK-GVKITLVNTISIWNKINNNIDLNSIETESISDG 199
Query: 71 FDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEF 130
+D GG++ A++ + Y + FW+VGP+SL+ L++ L + N+ VDC++YD+F+ W DV+K F
Sbjct: 200 YDNGGMSSAENMESYKDTFWKVGPKSLSQLLHKLQSSNNPVDCVVYDAFLNWTFDVSKSF 379
Query: 131 GIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQDPT 190
I A FLTQ +N+I +H + ++ P + EI LP LP LQ D+PSF + Y P
Sbjct: 380 EIPVAVFLTQACSVNTINFHAFMKWIELPISKSEIVLPGLPTLQDADLPSFLYQYGTYPG 559
Query: 191 FLDIGVAQFSNIHKADWILCNSFFELEKEVADW 223
+ DI QFS I + DW+L N+F+ELE V DW
Sbjct: 560 YFDILTNQFSKIDQVDWVLVNTFYELEPXVVDW 658
>TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyltransferase
HRA25 {Phaseolus vulgaris}, partial (47%)
Length = 1332
Score = 201 bits (511), Expect = 5e-52
Identities = 136/457 (29%), Positives = 224/457 (48%), Gaps = 15/457 (3%)
Frame = +1
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYK---NIPKLPNNSITIETIS 68
H LV+P+P GH NP+++ +L + G K+T + T ++K N + +I T+
Sbjct: 16 HFLVIPYPIPGHINPLMQLCHVLAKH-GCKITFLNTEFSHKRTNNNNEQSQETINFVTLP 192
Query: 69 DGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDH--VDCLIYDSFMPWCLDV 126
DG + + D K L + P L LI +NA +D + C+I M W L+V
Sbjct: 193 DGLEPED--DRSDQKKVLFSIKRNMPPLLPKLIEEVNALDDENKICCIIVTFNMGWALEV 366
Query: 127 AKEFGIVGASFLTQNLVMNSIYYHVHL----GKLKPPFV---EQEITL-PALPQLQPRDM 178
GI G T + + Y + G + + +QEI L P +P++ +++
Sbjct: 367 GHNLGIKGVLLWTGSATSLAFCYSIPKLIDDGVIDSAGIYTKDQEIQLSPNMPKMDTKNV 546
Query: 179 PSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCL 238
P F D D Q + W LCN+ ++LE T I F +GP +
Sbjct: 547 PWRTF----DKIIFDHLAQQMQTMKLGHWWLCNTTYDLEHA----TFSISPKFLPIGPLM 702
Query: 239 PYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCL 298
++D + S + S++WL+ +P +S VYVSFGS+A +++ Q E+A L
Sbjct: 703 ---------ENDSNKSSFWQEDMTSLDWLDKQPSQSVVYVSFGSLAVMDQNQFNELALGL 855
Query: 299 KDCGSYFLWVVKTSEETKLPKDF--EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNS 356
FLWVV+ S + K+ + E G +V+W PQ ++L H AI CF++HCGWNS
Sbjct: 856 DLLDKPFLWVVRPSNDNKVNYAYPDEFLGTKGKIVSWVPQKKILNHPAIACFISHCGWNS 1035
Query: 357 TLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSM 416
T+E + G+P + P +DQ + ++ D+WKVG DE IV K+ +K + +++
Sbjct: 1036TIEGVYSGIPFLCWPFATDQFTNKSYICDVWKVGFELDKDENGIVLKEEIKKKVEQLL-- 1209
Query: 417 SEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVN 453
+ ++I ++ K L + +DG S KN+ F+N
Sbjct: 1210--QDQDIKERSLKLKELTLENIVEDGKSSKNLQNFIN 1314
>TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltransferase-10
{Vigna angularis}, partial (41%)
Length = 1588
Score = 189 bits (480), Expect = 2e-48
Identities = 136/484 (28%), Positives = 226/484 (46%), Gaps = 39/484 (8%)
Frame = +2
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSIT-------I 64
H + +PFPA GH NP ++ +K+L + G +T V N+K + K +
Sbjct: 128 HVVCVPFPAQGHVNPFMQLAKILNKL-GFHITFVNNEFNHKRLIKSLGHDFLKGQPDFRF 304
Query: 65 ETISDGFDK---------GGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARND--HVDC 113
ETI DG GG+ E Y GP L L+ NLN ++ V
Sbjct: 305 ETIPDGLPPSDENATQSIGGLVEGCRKHCY-------GP--LKELVENLNNSSEVPQVTS 457
Query: 114 LIYDSFMPWCLDVAKEFGIVGASFLTQNL--VMNSIYYHVHLGKLKPPFVEQEITL---- 167
+IYD M + +DVAK+ GI F T + +M + + + + P+ ++
Sbjct: 458 IIYDGLMGFAVDVAKDLGIAEQQFWTASACGLMGYLQFDELIKRNMLPYKDESYITDGSL 637
Query: 168 -------PALPQLQPRDMPSFYFTYEQDP-TFLDIGVAQFSNIHKADWILCNSFFELEKE 219
P + ++ RD+PSF T D +F+ G+ + K+ I+ N+ ELE E
Sbjct: 638 DVHLDWTPGMKNIRMRDLPSFVRTTTLDEISFVGFGL-EAQQCMKSSAIIINTVKELESE 814
Query: 220 VADWTMKIWSNFRTVGPC--LPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVY 277
V D M I N +GP L F +K + S I+WL+ S +Y
Sbjct: 815 VLDALMAINPNIYNIGPLQFLANNFPEKENGFKSNGSSLWKNDLTCIKWLDQWEPSSVIY 994
Query: 278 VSFGSMASLNEEQIEEVAHCLKDCGSYFLWV-----VKTSEETKLPKDFEKKSENGLVVA 332
+++GS+A ++E+ E A L + FLW+ VK + ++ + G ++
Sbjct: 995 INYGSIAVMSEKHFNEFAWGLANSKLPFLWINRPDLVKGKCRPLSQEFLDEVKDRGYIIT 1174
Query: 333 WCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR 392
WCPQ EVLAH ++G F+THCGWNS+LE++S G P++ P +++Q + +++ W +G
Sbjct: 1175WCPQSEVLAHPSVGVFLTHCGWNSSLESISEGKPMIGWPFFAEQQTNCRYISTTWGIG-- 1348
Query: 393 PLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFV 452
+D K V+++ + + + E M EKGK+ ++WK A GSS+ + +
Sbjct: 1349--MDIKDDVKREEVTELVKE-MIKGEKGKKKREKCVEWKKKVVEAAKPGGSSYNDFYRLL 1519
Query: 453 NSLF 456
+
Sbjct: 1520KDAY 1531
>TC81622 weakly similar to PIR|T00584|T00584 indole-3-acetate
beta-glucosyltransferase homolog T27E13.12 - Arabidopsis
thaliana, partial (25%)
Length = 1735
Score = 189 bits (480), Expect = 2e-48
Identities = 131/449 (29%), Positives = 223/449 (49%), Gaps = 14/449 (3%)
Frame = +2
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQE--GVKVTLVTTISNYKNIPKLPN-NSITIETIS 68
H + +PFP GH NPML F K+L Q+ + +T V T I P SI TI
Sbjct: 167 HVVAMPFPGRGHINPMLSFCKILTSQKPNNLLITFVLTEEWLTFIGADPKPESIRFATIP 346
Query: 69 DGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAK 128
+ +A DF + L++ L VD ++ D + W ++V
Sbjct: 347 NVIPPER-EKAGDFPGFYEAVMTKMEAPFEKLLDQLELP---VDVIVGDVELRWPVNVGN 514
Query: 129 EFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFV-----EQEITLPALPQLQPRDMPSFYF 183
+ A+F T + S+ +H+ + K EQ +P + D+ +
Sbjct: 515 RRNVPVAAFWTMSASFYSMLHHLDVFSRKHHLTVDKLDEQAENIPGISSFHIEDVQTVLC 694
Query: 184 TYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWS-NFRTVGPCLPYTF 242
+ D L + + S + KA+++L + ELE E D I+ +GP +PY
Sbjct: 695 --KNDHQVLQLALGCISKVPKANYLLLTTVQELEAETIDSLKSIFPFPIYPIGPSIPYLD 868
Query: 243 LDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCG 302
++++ + DHS + I+WL+++P S +Y+S GS S++ Q++E+ L + G
Sbjct: 869 IEEKNPANTDHS------QDYIKWLDSQPSESVLYISLGSFLSVSNAQMDEIVEALNNSG 1030
Query: 303 SYFLWVVKTSEETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALS 362
+L+V + ET KD K + G+V+ WC QL+VL+H +IG F +HCGWNSTLE +
Sbjct: 1031IRYLYVARG--ETSRLKD--KCGDKGMVIPWCDQLKVLSHSSIGGFWSHCGWNSTLETVF 1198
Query: 363 IGVPIVAIPLYSDQGIDAKFLVDIWKVG----IRPLVDEKQIVRKDPLKDCICEIMSM-S 417
GVPI+ PL+ DQ ++ +VD WK G I+ ++ I+ K+ +++ + M + +
Sbjct: 1199AGVPILTFPLFLDQVPNSTQIVDEWKNGWKVEIQSKLESDVILAKEDIEELVKRFMDLEN 1378
Query: 418 EKGKEIMNNVMQWKTLATRAVGKDGSSHK 446
++GK+I + + K + +A+GK GSS +
Sbjct: 1379QEGKKIRDRARELKVMFRKAIGKGGSSDR 1465
>TC78695 similar to GP|4115563|dbj|BAA36423.1 UDP-glucose:anthocyanin
5-O-glucosyltransferase {Verbena x hybrida}, partial
(49%)
Length = 1741
Score = 188 bits (478), Expect = 3e-48
Identities = 96/233 (41%), Positives = 151/233 (64%), Gaps = 19/233 (8%)
Frame = +1
Query: 242 FLDKRVKDDEDHS----IAQLKS-DESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAH 296
FLD KD D+S + ++ S D+ I+WL++K ++S VYVSFG++A L++ Q+EE+A
Sbjct: 802 FLDG--KDPTDNSFGGDVVRVDSKDDYIQWLDSKDEKSVVYVSFGTLAVLSKRQMEEIAR 975
Query: 297 CLKDCGSYFLWVVKTSEETKLPKDFEKKSE--------------NGLVVAWCPQLEVLAH 342
L D G FLWV++ + KL + E++ + NG +V WC Q+EVL+H
Sbjct: 976 ALLDSGFSFLWVIR---DKKLQQQKEEEVDDDELSCREELENNMNGKIVKWCSQVEVLSH 1146
Query: 343 EAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVR 402
++GCF+THCGWNSTLE+L GVP+VA P ++DQ +AK + D+WK G+R DE+ +V+
Sbjct: 1147 RSLGCFMTHCGWNSTLESLGSGVPMVAFPQWTDQTTNAKLIEDVWKTGLRMEHDEEGMVK 1326
Query: 403 KDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+ ++ C+ +M EKG+E+ N +WK LA AV + GSS++N+ ++N +
Sbjct: 1327 VEEIRKCLEVVMGKGEKGEELRRNAKKWKDLARAAVKEGGSSNRNLRSYLNDI 1485
Score = 80.1 bits (196), Expect = 2e-15
Identities = 59/190 (31%), Positives = 92/190 (48%), Gaps = 10/190 (5%)
Frame = +2
Query: 2 ENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYK---NIPKLP 58
++ T ++ H L++ +P GH NP L+F+K L G KVT TTI Y N P +P
Sbjct: 53 QSTTTMAQNHHFLIITYPLQGHINPALQFTKRLISL-GAKVTFATTIHLYSRLINKPTIP 229
Query: 59 NNSITIETISDGFDKGGVAEA-KDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYD 117
++ T SDG+D G + +D Y+++F + G + L ++I + N CLIY
Sbjct: 230 G--LSFATFSDGYDDGQKSFGDEDIVSYMSEFTRRGSEFLTNIILSSKQENHPFTCLIYT 403
Query: 118 SFMPWCLDVAKEFGIVGASFLTQNLVMNSI---YYHVHLGKLKPPFVEQE--ITLPALP- 171
+ W VA E + Q + I Y+H H + ++ I+LP L
Sbjct: 404 LILSWAPKVAHELHLPSTLLWIQAATVFDIFYYYFHEHGDYITNKSKDETCLISLPGLSF 583
Query: 172 QLQPRDMPSF 181
L+ RD+PSF
Sbjct: 584 SLKSRDLPSF 613
>TC91517 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (31%)
Length = 1027
Score = 113 bits (283), Expect(2) = 6e-45
Identities = 49/118 (41%), Positives = 81/118 (68%)
Frame = +1
Query: 338 EVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDE 397
+VLAH ++ CF+THCGWNS++EA+++GVP++ P + DQ +AKFLVD++ GIR +
Sbjct: 532 QVLAHSSVACFITHCGWNSSMEAITLGVPMLTFPAFGDQLTNAKFLVDVFGAGIRLGYGD 711
Query: 398 KQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
K++V +D +K C+ E ++ EK + + N M+WKT A A+ GSS +++ F+ +
Sbjct: 712 KKLVTRDEVKKCLLEAIT-GEKAERLKQNAMKWKTAAEDAMAVGGSSDRHLDAFIQDI 882
Score = 85.9 bits (211), Expect(2) = 6e-45
Identities = 59/187 (31%), Positives = 97/187 (51%), Gaps = 8/187 (4%)
Frame = +2
Query: 162 EQEITLPALPQ---LQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEK 218
++E+TL +L L+ ++P + + P + AQ ++ K +L ++F ELE
Sbjct: 2 DEELTLMSLNSSIVLKYNEIPDYIHPFNPFPILGTLTTAQIKDMSKVFCVLVDTFEELEH 181
Query: 219 EVADWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQL--KSDES--IEWLNNKPKRS 274
+ D+ K R VGP F + + ++++ +D+S IEWLN KPK S
Sbjct: 182 DFIDYISKKSITIRHVGPL----FKNPKANGASNNTLGDFTKSNDDSTIIEWLNTKPKGS 349
Query: 275 AVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDF-EKKSENGLVVAW 333
VY+SFG++ + +EQ+ E+A+ L + FLWV+K + P F E+ S G VV W
Sbjct: 350 VVYISFGTIVNHTQEQVNEIAYGLLNSQVSFLWVLK---QHVFPDGFLEETSGRGKVVKW 520
Query: 334 CPQLEVL 340
PQ + L
Sbjct: 521 SPQNKYL 541
>TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase-8 {Vigna
angularis}, partial (53%)
Length = 1787
Score = 177 bits (450), Expect = 6e-45
Identities = 142/496 (28%), Positives = 238/496 (47%), Gaps = 45/496 (9%)
Frame = +1
Query: 5 TISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPK-LPNNSIT 63
T + +H + PF A+GH P ++ +++ + G+KVT+VTT N I + + I
Sbjct: 37 TNENQELHIIFFPFLANGHIIPCVDLARVFSSR-GLKVTIVTTHLNVPLISRTIGKAKIN 213
Query: 64 IETIS----------DGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDC 113
I+TI +G + A A D + K + + L H++ + DC
Sbjct: 214 IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQ-----EKPDC 378
Query: 114 LIYDSFMPWCLDVAKEFGIV-----GASFLTQNLVMNSIYYHVH--LGKLKPPFV----- 161
L+ D F PW D A +F I G F ++ + Y + PFV
Sbjct: 379 LVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLP 558
Query: 162 -EQEITLPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEV 220
E +T LPQL + D F + + K+ ++ N+F+ELE
Sbjct: 559 GEITLTKMQLPQLP-----------QHDKVFTKLLEESNESEVKSFGVIANTFYELEPVY 705
Query: 221 AD-WTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSI----AQLKSDESIEWLNNKPKRSA 275
AD + ++ +GP + +D E+ + A + E ++WL +K S
Sbjct: 706 ADHYRNELGRKAWHLGP------VSLCNRDTEEKACRGREASIDEHECLKWLQSKEPNSV 867
Query: 276 VYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETK------LPKDFEKKSEN-- 327
+YV FGSM ++ Q++E+A L+ F+WVV+ S +++ LP+ FE++ E
Sbjct: 868 IYVCFGSMTVFSDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSG 1047
Query: 328 -GLVV-AWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVD 385
GL++ W PQ+ +L HE++G FVTHCGWNSTLE +S G+P+V P+Y +Q +AKFL D
Sbjct: 1048KGLIIRGWAPQVMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSD 1227
Query: 386 IWKVGIRPLVDE------KQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVG 439
I K+G+ V + V+KD ++ + IM + ++ +E+ + ++ +A RAV
Sbjct: 1228IVKIGVGVGVQTWIGMGGGEPVKKDVIEKAVRRIM-VGDEAEEMRSRAKEFGKMARRAVE 1404
Query: 440 KDGSSHKNMIEFVNSL 455
GSS+ + + L
Sbjct: 1405VGGSSYNDFSNLIEDL 1452
>TC91074 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (35%)
Length = 911
Score = 152 bits (385), Expect(2) = 2e-44
Identities = 79/179 (44%), Positives = 115/179 (64%), Gaps = 9/179 (5%)
Frame = +3
Query: 286 LNEEQIEEVAHCLKDCGSYFLWVVKTS------EETKLPKDF-EKKSENGLVVAWCPQLE 338
L +EQ+ E+AH L D LWV+K +E LP +F E+ +E G VV W PQ E
Sbjct: 249 LPQEQVNEIAHGLLDSNVSLLWVLKPPSKESGRKEHVLPNEFLEETNERGKVVNWSPQEE 428
Query: 339 VLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVD-- 396
VLAH ++ CF+THCGWNS++EALS+GVP++ P + DQ +AKFLVD++ VGIR
Sbjct: 429 VLAHPSVACFITHCGWNSSMEALSLGVPMLTFPAWGDQVTNAKFLVDVFGVGIRLGYSHA 608
Query: 397 EKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+ ++V +D +K C+ E ++ EKG+E+ N ++WK A AV GSS +N+ EF+ +
Sbjct: 609 DNKLVTRDEVKKCLLE-ATIGEKGEELKQNAIKWKKAAEEAVATGGSSDRNLDEFMEDI 782
Score = 44.7 bits (104), Expect(2) = 2e-44
Identities = 30/84 (35%), Positives = 45/84 (52%), Gaps = 2/84 (2%)
Frame = +2
Query: 202 IHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSD 261
+ K +L +SF ELE + D+ K R +GP F + ++K D +KSD
Sbjct: 2 LSKVFCVLVDSFDELEHDYIDYISKKSILTRPIGPL----FNNPKIKCASDIRGDFVKSD 169
Query: 262 ES--IEWLNNKPKRSAVYVSFGSM 283
+ IEWLN+K S VY+SFG++
Sbjct: 170 DCNIIEWLNSKANDSVVYISFGTI 241
>TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (76%)
Length = 1643
Score = 174 bits (440), Expect = 8e-44
Identities = 141/484 (29%), Positives = 237/484 (48%), Gaps = 35/484 (7%)
Frame = +3
Query: 5 TISTKSVHCLVLPFPAHGHTNPMLEFSK-LLQQQEGVKVTLVTTI------SNYKNIPKL 57
T K++H V+P P H P++EFSK L+ VT + S+ + K+
Sbjct: 15 TKMAKTIHIAVIPSPGFSHLVPIVEFSKRLVTNHPNFHVTCIIPSLGSPPDSSKSYLEKI 194
Query: 58 PNN--SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLI 115
P N SI + I+ GV A + + + + S+ + +L+++ V +I
Sbjct: 195 PPNINSIFLPPINKQDLPQGVYPA----ILIQQTVTLSLPSIHQALKSLSSKAPLV-AII 359
Query: 116 YDSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPF------VEQEITLPA 169
DSF LD AKEF + + + N + + +HL KL +++ I L
Sbjct: 360 ADSFAFEALDFAKEFNSLSYFYFPTSA--NVLSFILHLPKLDEEVSCEFKDLQEPIKLQG 533
Query: 170 LPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVAD-WTMKIW 228
+ D+P+ T ++ + + + + + D IL NSF+ELE + K +
Sbjct: 534 CVPINGIDLPTP--TKDRSSEAYRMFLQRAKSFYFVDGILINSFYELESSAVEALKQKGY 707
Query: 229 SN--FRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASL 286
N + VGP + V DE E ++WL N+P+ S +YVSFGS +L
Sbjct: 708 GNISYFPVGPITQIGSSNNDVVGDEH---------ECLKWLKNQPQNSVLYVSFGSGGTL 860
Query: 287 NEEQIEEVAHCLKDCGSYFLWVVKTSEETK---------------LPKDFEKKS-ENGLV 330
++ QI E+A L+ G F+W V+ ++ LP+ F++++ E G +
Sbjct: 861 SQRQINEIAFGLELSGQRFIWGVRAPSDSVNAAYLESTNEDPLKFLPEGFQERTKEKGFI 1040
Query: 331 V-AWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKV 389
+ +W PQ+E+L H ++G F++HCGWNS LE++ GVPIVA PL+++Q ++A L D KV
Sbjct: 1041LPSWAPQVEILKHSSVGGFLSHCGWNSVLESMQEGVPIVAWPLFAEQAMNAVMLCDGLKV 1220
Query: 390 GIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMI 449
IR ++ +IV KD + I +M E+GK + + + K A AV +GSS +N+
Sbjct: 1221AIRLKFEDDEIVEKDKTANVIKCLME-GEEGKTMRDRMKSLKDYAVNAVKDEGSSIQNLS 1397
Query: 450 EFVN 453
+ +
Sbjct: 1398QLAS 1409
>TC86394 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyltransferase
HRA25 {Phaseolus vulgaris}, partial (34%)
Length = 1517
Score = 109 bits (273), Expect(2) = 9e-42
Identities = 55/172 (31%), Positives = 97/172 (55%), Gaps = 2/172 (1%)
Frame = +2
Query: 284 ASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDF--EKKSENGLVVAWCPQLEVLA 341
A +++ Q E+A L F+WVV+ S + K+ + E G +V W PQ ++L
Sbjct: 890 AVMDQNQFNELALGLDLLDKPFIWVVRPSNDNKVNYAYPDEFLGTKGKIVGWAPQKKILN 1069
Query: 342 HEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIV 401
H AI CF++HCGWNST+E + GVP + P + DQ ++ ++ D+WKVG+ DE ++
Sbjct: 1070 HPAIACFISHCGWNSTVEGVYSGVPFLCWPFHGDQFMNKSYVCDVWKVGLELDKDEDGLL 1249
Query: 402 RKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVN 453
K ++ + +++ ++I ++ K L + + ++G S KN+I F+N
Sbjct: 1250 PKREIRIKVEQLLG----DQDIKERSLKLKDLTLKNIVENGHSSKNLINFIN 1393
Score = 79.0 bits (193), Expect(2) = 9e-42
Identities = 75/294 (25%), Positives = 122/294 (40%), Gaps = 25/294 (8%)
Frame = +3
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYK-------NIPKLPN----- 59
H LV+P+P GH NP+++ S LL + G K+T + T + K +I K N
Sbjct: 60 HFLVIPYPIAGHVNPLMQLSHLL-SKHGCKITFLNTEFSNKRTNKNNISISKKDNLKNEQ 236
Query: 60 --NSITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARN--DHVDCLI 115
+I T+ DG + D + + + P L +LI ++NA + + + C+I
Sbjct: 237 SQETINFVTLPDGLEDED--NRSDQRKVIFSIRRNMPPLLPNLIEDVNAMDAENKISCII 410
Query: 116 YDSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVH-------LGKLKPPFVEQEITL- 167
M W L+V GI G T + + Y + + P +QEI L
Sbjct: 411 VTFNMGWALEVGHSLGIKGVLLWTASATSLAYCYSIPKLIDDGVMDSAGIPTTKQEIQLF 590
Query: 168 PALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKI 227
P +P + + P D D + + DW LCN+ + LE T I
Sbjct: 591 PNMPMIDTANFP----WRAHDKILFDYISQEMQAMKFGDWWLCNTTYNLEHA----TFSI 746
Query: 228 WSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDES-IEWLNNKPKRSAVYVSF 280
F +GP + ED++ + + D + ++WL+ P +S YVSF
Sbjct: 747 SPKFLPIGPFMSI----------EDNTSSFWQEDATCLDWLDQYPPQSVAYVSF 878
>AW684227 similar to GP|7385017|gb|A UDP-glucose:salicylic acid
glucosyltransferase {Nicotiana tabacum}, partial (7%)
Length = 630
Score = 150 bits (380), Expect(2) = 3e-41
Identities = 76/124 (61%), Positives = 91/124 (73%)
Frame = +3
Query: 1 MENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNN 60
ME K I+ K VHCLVLP+PA GH NPML+FSK LQ EG++VTLVTT+ + K + +P
Sbjct: 69 MEKKVITNK-VHCLVLPYPAQGHINPMLQFSKDLQH-EGIRVTLVTTLYHRKTLQSVPP- 239
Query: 61 SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFM 120
S TIETISDGFD GGV EA +K YL +FWQVGP++LA LI + D VDC+IYDSF
Sbjct: 240 SFTIETISDGFDNGGVEEAGGYKAYLGRFWQVGPKTLAQLIEKFGSLGDKVDCVIYDSFF 419
Query: 121 PWCL 124
PW L
Sbjct: 420 PWAL 431
Score = 36.2 bits (82), Expect(2) = 3e-41
Identities = 19/34 (55%), Positives = 23/34 (66%), Gaps = 1/34 (2%)
Frame = +2
Query: 141 NLVMNSIYYHVHLGKLKPPFVEQEI-TLPALPQL 173
N+ +NSIYYHVHL K K P +E I T LP+L
Sbjct: 479 NMSVNSIYYHVHLKK*KVPLIEDXIFTFLWLPKL 580
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,054,822
Number of Sequences: 36976
Number of extensions: 239282
Number of successful extensions: 1586
Number of sequences better than 10.0: 188
Number of HSP's better than 10.0 without gapping: 1409
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1421
length of query: 458
length of database: 9,014,727
effective HSP length: 99
effective length of query: 359
effective length of database: 5,354,103
effective search space: 1922122977
effective search space used: 1922122977
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142095.2


