

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.7 - phase: 0
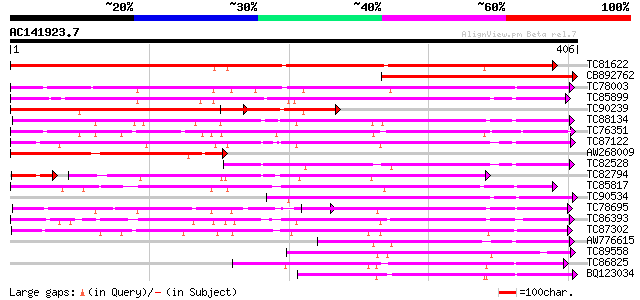
(406 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81622 weakly similar to PIR|T00584|T00584 indole-3-acetate bet... 380 e-106
CB892762 weakly similar to GP|21536917|gb| putative glucosyltran... 295 2e-80
TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase... 199 2e-51
TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:... 182 2e-46
TC90239 weakly similar to PIR|T00583|T00583 probable indole-3-ac... 174 6e-44
TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid UDP-gluco... 167 9e-42
TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycos... 153 1e-37
TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltran... 151 4e-37
AW268009 weakly similar to PIR|T00583|T005 probable indole-3-ace... 146 2e-35
TC82528 weakly similar to PIR|C86356|C86356 hypothetical protein... 144 6e-35
TC82794 weakly similar to GP|3928543|dbj|BAA34687.1 UDP-glucose ... 130 1e-34
TC85817 weakly similar to PIR|B85014|B85014 probable flavonol gl... 143 1e-34
TC90534 weakly similar to GP|19911197|dbj|BAB86925. glucosyltran... 137 1e-32
TC78695 similar to GP|4115563|dbj|BAA36423.1 UDP-glucose:anthocy... 135 4e-32
TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyl... 134 8e-32
TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase... 131 4e-31
AW776615 weakly similar to GP|14532546|gb At1g22370/T16E15_3 {Ar... 128 5e-30
TC89558 similar to GP|14349253|dbj|BAB60721. glucosyltransferase... 124 9e-29
TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransfe... 123 1e-28
BQ123034 weakly similar to GP|14334982|gb putative glucosyltrans... 123 1e-28
>TC81622 weakly similar to PIR|T00584|T00584 indole-3-acetate
beta-glucosyltransferase homolog T27E13.12 - Arabidopsis
thaliana, partial (25%)
Length = 1735
Score = 380 bits (975), Expect = e-106
Identities = 202/431 (46%), Positives = 275/431 (62%), Gaps = 39/431 (9%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNN-SNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIP 59
MPFP RG+INPM++ CK+L S +N+ ++FV+TEEWL+FI ++PKP++I F + NVIP
Sbjct: 179 MPFPGRGHINPMLSFCKILTSQKPNNLLITFVLTEEWLTFIGADPKPESIRFATIPNVIP 358
Query: 60 SELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRNIPAAL 119
E D P F E VMTKMEAPFE+LLD L+ P +IV D L W V V NRRN+P A
Sbjct: 359 PEREKAGDFPGFYEAVMTKMEAPFEKLLDQLELPVDVIVGDVELRWPVNVGNRRNVPVAA 538
Query: 120 FWPMPASIFSVFLHQHIFEQNGHYPV--------KYPGFEWIH----------------- 154
FW M AS +S+ H +F + H V PG H
Sbjct: 539 FWTMSASFYSMLHHLDVFSRKHHLTVDKLDEQAENIPGISSFHIEDVQTVLCKNDHQVLQ 718
Query: 155 ----------KAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSN 204
KA YLL +++ ELE++ ID LKS P PIY IGP+IP + + P+N
Sbjct: 719 LALGCISKVPKANYLLLTTVQELEAETIDSLKSIFPFPIYPIGPSIPYLDI--EEKNPAN 892
Query: 205 TNHSY-YIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRL 263
T+HS YI+WLDSQP SVLYI+ GSF SVS+AQ+DEI AL S +R+L++AR E SRL
Sbjct: 893 TDHSQDYIKWLDSQPSESVLYISLGSFLSVSNAQMDEIVEALNNSGIRYLYVARGETSRL 1072
Query: 264 KEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPF 323
K+ CG G+++ WCDQL+VLSH SIGGFWSHCGWNST E++ AGVP LT P+++DQ
Sbjct: 1073KDKCG--DKGMVIPWCDQLKVLSHSSIGGFWSHCGWNSTLETVFAGVPILTFPLFLDQVP 1246
Query: 324 NSKMMVEDWKVGCRV--KEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKI 381
NS +V++WK G +V + ++ D ++ K+ I +LV FMDL+ + + IR+R+++L+ +
Sbjct: 1247NSTQIVDEWKNGWKVEIQSKLESDVILAKEDIEELVKRFMDLENQEGKKIRDRARELKVM 1426
Query: 382 CLNSIANGGSA 392
+I GGS+
Sbjct: 1427FRKAIGKGGSS 1459
>CB892762 weakly similar to GP|21536917|gb| putative glucosyltransferase
{Arabidopsis thaliana}, partial (15%)
Length = 572
Score = 295 bits (755), Expect = 2e-80
Identities = 139/140 (99%), Positives = 140/140 (99%)
Frame = +2
Query: 267 CGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSK 326
CGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSK
Sbjct: 2 CGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSK 181
Query: 327 MMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSI 386
MMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKL+KICLNSI
Sbjct: 182 MMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLKKICLNSI 361
Query: 387 ANGGSAHTDFNAFISDVMHL 406
ANGGSAHTDFNAFISDVMHL
Sbjct: 362 ANGGSAHTDFNAFISDVMHL 421
>TC78003 similar to GP|20146093|dbj|BAB88935. glucosyltransferase NTGT2
{Nicotiana tabacum}, partial (50%)
Length = 1740
Score = 199 bits (505), Expect = 2e-51
Identities = 136/464 (29%), Positives = 225/464 (48%), Gaps = 60/464 (12%)
Frame = +3
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P+PV+G+INP K L++ + HV+ T + I+++P N+S+ S+
Sbjct: 162 VPYPVQGHINPAFEFAKRLITLGA--HVTISTTVHMHNRITNKPTLPNLSYYPFSDGY-D 332
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLL------DHPPSIIVYDTLLYWAVVVANRRN 114
+ G A++E E + D++ P + +V+ LL WA A +
Sbjct: 333 DGFKGTGSDAYLEYHAEFQRRGSEFVSDIILKNSQEGTPFTCLVHSLLLQWAAEAAREFH 512
Query: 115 IPAALFWPMPASIFSV-FLHQHIFEQNGHYP---VKYPGFEWIHKAQ------------- 157
+P AL W PA++F + + + H F + P ++ PG + ++
Sbjct: 513 LPTALLWVQPATVFDILYYYFHGFSDSIKNPSSSIELPGLPLLFSSRDLPSFLLASCPDA 692
Query: 158 YLLFSSIYELESQAIDV--------------------LKSKLPLPIYTIGPTIPKFSLIK 197
Y L +S +E + +DV L++ + +IGP IP L
Sbjct: 693 YSLMTSFFEEQFNELDVETNLTKTILVNSFESLEPKALRAVKKFNMISIGPLIPSEHL-- 866
Query: 198 NDPKPSNTNHSY------------YIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAAL 245
D K S ++SY +EWLDS+P SV+Y++ GS+F +S Q +EIA AL
Sbjct: 867 -DEKDSTEDNSYGGQTHIFQPSNDCVEWLDSKPKSSVVYVSFGSYFVLSERQREEIAHAL 1043
Query: 246 CASNVRFLWIARSEASRLKEICGAHH-----MGLIMEWCDQLRVLSHPSIGGFWSHCGWN 300
FLW+ R + E + G I++WC Q+ +LSHPS+G F +HCGWN
Sbjct: 1044LDCGFPFLWVLREKEGENNEEGFKYREELEEKGKIVKWCSQMEILSHPSLGCFLTHCGWN 1223
Query: 301 STKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEF 360
ST ESLV GVP + P + DQ N+K++ + WK+G RV E+V D +V+ D+I + +
Sbjct: 1224STLESLVKGVPMVAFPQWTDQMTNAKLIEDVWKIGVRVDEEVNEDGIVRGDEIRRCLEVV 1403
Query: 361 MDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
M GE ++R KK +++ ++ GGS+ + +F+ V+
Sbjct: 1404MG-SGEKGEELRRSGKKWKELAREAVKEGGSSEKNLRSFLDGVV 1532
>TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:salicylic
acid glucosyltransferase {Nicotiana tabacum}, partial
(47%)
Length = 1617
Score = 182 bits (462), Expect = 2e-46
Identities = 131/446 (29%), Positives = 210/446 (46%), Gaps = 45/446 (10%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P+P +G++NPM+ K L+ I + VT W + S +I S S+
Sbjct: 56 LPYPAQGHMNPMIQFSKRLIEKGVKITL-ITVTSFWK--VISNKNLTSIDVESISDGYDE 226
Query: 61 E-LICGRDHPAFMEDVMTKMEAPFEELLDLL---DHPPSIIVYDTLLYWAVVVANRRNIP 116
L+ + E ELL L ++PP+ +++D L W + V +
Sbjct: 227 GGLLAAESLEDYKETFWKVGSQTLSELLHKLSSSENPPNCVIFDAFLPWVLDVGKSFGLV 406
Query: 117 AALFWPMPASIFSVFLHQH----------------------------IFEQNGHYP---- 144
F+ S+ SV+ H H + G YP
Sbjct: 407 GVAFFTQSCSVNSVYYHTHEKLIELPLTQSEYLLPGLPKLAPGDLPSFLYKYGSYPGYFD 586
Query: 145 VKYPGFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKN--DPKP 202
+ F I KA ++L +SIYELE + +D L PL TIGP++P L K D K
Sbjct: 587 IVVNQFANIGKADWILANSIYELEPEVVDWLVKIWPLK--TIGPSVPSMLLDKRLKDDKE 760
Query: 203 -----SNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR 257
S+ N + I+WL+ +P GSV+Y + GS +S Q E+A L S FLW+ R
Sbjct: 761 YGVSLSDPNTEFCIKWLNDKPKGSVVYASFGSMAGLSEEQTQELALGLKDSESYFLWVVR 940
Query: 258 S-EASRL-KEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTL 315
+ S+L K + GLI+ WC QL VL+H ++G F +HCGWNST E+L GVP + +
Sbjct: 941 ECDQSKLPKGFVESSKKGLIVTWCPQLLVLTHEALGCFVTHCGWNSTLEALSIGVPLIAM 1120
Query: 316 PIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERS 375
P++ DQ N+K++ + WK+G R D K +V+ + I + E ++ E +I++ +
Sbjct: 1121PLWTDQVTNAKLIADVWKMGVRAVADEKE--IVRSETIKNCIKEI--IETEKGNEIKKNA 1288
Query: 376 KKLQKICLNSIANGGSAHTDFNAFIS 401
K + + +S+ GG + + F++
Sbjct: 1289LKWKNLAKSSVDEGGRSDKNIEEFVA 1366
>TC90239 weakly similar to PIR|T00583|T00583 probable indole-3-acetate
beta-glucosyltransferase T27E13.11 - Arabidopsis
thaliana, partial (30%)
Length = 916
Score = 174 bits (441), Expect = 6e-44
Identities = 89/174 (51%), Positives = 118/174 (67%), Gaps = 3/174 (1%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSE-PKPDN--ISFRSGSNV 57
+P+P RG+INPMMNLCKLL+SNN NI V+FVVTEEWL+ I+S+ PKP+N I F + NV
Sbjct: 68 VPYPGRGHINPMMNLCKLLISNNPNIVVTFVVTEEWLTIINSDSPKPNNKNIKFATIPNV 247
Query: 58 IPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRNIPA 117
IPSE G+D F+E V+TKME PFE+LLD L+ P++I++D+ L+W + VAN+RN+P
Sbjct: 248 IPSEEGRGKDFLNFLEAVVTKMEDPFEKLLDSLETAPNVIIHDSYLFWVIRVANKRNVPV 427
Query: 118 ALFWPMPASIFSVFLHQHIFEQNGHYPVKYPGFEWIHKAQYLLFSSIYELESQA 171
A FWPM AS F V H E++GHYP + + S I+ QA
Sbjct: 428 ASFWPMSASFFLVLKHYRRLEEHGHYPKLVTNVWITFRGTHQFVSKIFHFMMQA 589
Score = 106 bits (264), Expect = 2e-23
Identities = 55/89 (61%), Positives = 65/89 (72%), Gaps = 3/89 (3%)
Frame = +1
Query: 152 WIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSY-- 209
W+ KAQYLLF SIYE+E QAIDVL+ + +PIYTIGPTIP FS N +TN
Sbjct: 634 WMKKAQYLLFPSIYEIEPQAIDVLREEFSIPIYTIGPTIPYFS--HNQIASLSTNQDVEL 807
Query: 210 -YIEWLDSQPIGSVLYIAQGSFFSVSSAQ 237
YI WLD+QPIGSVLY++QGSF +VSS Q
Sbjct: 808 DYINWLDNQPIGSVLYVSQGSFLTVSSEQ 894
>TC88134 weakly similar to SP|Q9MB73|LGT_CITUN Limonoid
UDP-glucosyltransferase (EC 2.4.1.210) (Limonoid
glucosyltransferase) (Limonoid GTase), partial (27%)
Length = 1669
Score = 167 bits (422), Expect = 9e-42
Identities = 125/462 (27%), Positives = 219/462 (47%), Gaps = 60/462 (12%)
Frame = +2
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS-- 60
F +G+INP++ L K L++ N+ ++ F S+ D + +N I
Sbjct: 92 FSAQGHINPLLRLGKSLLTKGLNVTLATTELVYHRVFKSTTTDTDTVPTSYSTNGINVLF 271
Query: 61 -----ELICGRDHP-AFMEDVMTKMEAPFEELLD--LLDHPP---SIIVYDTLLYWAVVV 109
++ G P +ME + L++ +++P + I+ + + W V
Sbjct: 272 FSDGFDISQGHKSPDEYMELIAKFGPISLTNLINNNFINNPSKKLACIINNPFVPWVANV 451
Query: 110 ANRRNIPAALFWPMPASIFSVF------LHQHIFEQNGHYPVKYPG-------------- 149
A NIP A W P +++S++ L+Q +N V+ PG
Sbjct: 452 AYELNIPCACLWIQPCTLYSIYYRFYNELNQFPTLENPEIDVELPGLPLLKPQDLPSFIL 631
Query: 150 ---------------FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFS 194
F+ + K +++L +S YELE ID + P I +GP +P S
Sbjct: 632 PSNPIKTMSDVLAEMFQDMKKLKWVLANSFYELEKDVIDSMAEIFP--ITPVGPLVPP-S 802
Query: 195 LIKNDPKPSN-----TNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASN 249
L+ D K T +EWL+ + SV+YI+ GS +S Q+ IA+AL SN
Sbjct: 803 LLGQDQKQDVGIEMWTPQDSCMEWLNQKLPSSVIYISFGSLIFLSEKQMQSIASALKNSN 982
Query: 250 VRFLWIARS-----EASRLKE--ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNST 302
FLW+ +S E +L E + GL++ WC Q +VL HP+I F +HCGWNST
Sbjct: 983 KYFLWVMKSNDIGAEKVQLSEKFLEETKEKGLVVTWCPQTKVLVHPAIACFLTHCGWNST 1162
Query: 303 KESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMD 362
E++ AGVP + P + DQP N+ ++ + +++G R+K+D D V+ D++ + + E
Sbjct: 1163LEAITAGVPMIGYPQWTDQPTNATLVSDVFRMGIRLKQD--NDGFVESDEVERAIEEI-- 1330
Query: 363 LDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
+ GE + ++ + +L+ ++A+GGS+ + F+ +++
Sbjct: 1331VGGERSEVFKKNALELKHAAREAVADGGSSDRNIQRFVDEIL 1456
>TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycose {Cicer
arietinum}, partial (92%)
Length = 1645
Score = 153 bits (386), Expect = 1e-37
Identities = 127/474 (26%), Positives = 222/474 (46%), Gaps = 69/474 (14%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDN----------IS 50
+PF +G++ P++NL +L+ S N HV+ + T + + I
Sbjct: 110 LPFFAQGHLIPLVNLARLVASKNQ--HVTIITTPSNAQLFDKTIEEEKAAGHHIRVHIIK 283
Query: 51 FRSGSNVIPS---ELICGRDHPAFMEDVMTK--MEAPFEELLDLLDHPPSIIVYDTLLYW 105
F S +P+ L D+ + M ++A EE + ++PP + + D + W
Sbjct: 284 FPSAQLGLPTGVENLFAASDNQTAGKIHMAAHFVKADIEEFMK--ENPPDVFISDIIFTW 457
Query: 106 AVVVANRRNIPAALFWPMPASIFSVFLHQHI-------FEQNGHY-----------PVK- 146
+ A IP +F P+ SIF V + Q I +G Y P+K
Sbjct: 458 SESTAKNLQIPRLVFNPI--SIFDVCMIQAIQSHPESFVSDSGPYQIHGLPHPLTLPIKP 631
Query: 147 YPGF--------EWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPT-----IPKF 193
PGF E + + ++ +S EL+ + ++ ++ +GPT IPK
Sbjct: 632 SPGFARLTESLIEAENDSHGVIVNSFAELDEGYTEYYENLTGRKVWHVGPTSLMVEIPKK 811
Query: 194 SLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
+ + S+ + WLD++ SVLYI+ GS +S+ Q+ E+A + AS +FL
Sbjct: 812 KKVVSTENDSSITKHQSLTWLDTKEPSSVLYISFGSLCRLSNEQLKEMANGIEASKHQFL 991
Query: 254 WIARSE-------------ASRLKEICGAHHMGLIME-WCDQLRVLSHPSIGGFWSHCGW 299
W+ + R+KE G++++ W Q +L HPSIGGF +HCGW
Sbjct: 992 WVVHGKEGEDEDNWLPKGFVERMKE----EKKGMLIKGWVPQALILDHPSIGGFLTHCGW 1159
Query: 300 NSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRV--------KEDVKRDTLVKKD 351
N+T E++ +GVP +T+P + DQ +N K++ E ++G V D K+ T+V+ +
Sbjct: 1160NATVEAISSGVPMVTMPGFGDQYYNEKLVTEVHRIGVEVGAAEWSMSPYDAKK-TVVRAE 1336
Query: 352 KIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMH 405
+I K V + MD +GE +IR+R+K++++ ++ GGS+ + D +H
Sbjct: 1337RIEKAVKKLMDSNGE-GGEIRKRAKEMKEKAWKAVQEGGSSQNCLTKLV-DYLH 1492
>TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltransferase-10
{Vigna angularis}, partial (41%)
Length = 1588
Score = 151 bits (382), Expect = 4e-37
Identities = 130/479 (27%), Positives = 203/479 (42%), Gaps = 74/479 (15%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTE-------EWLSFISSEPKPDNISFRS 53
+PFP +G++NP M L K+L N H++FV E + L + +PD
Sbjct: 140 VPFPAQGHVNPFMQLAKIL--NKLGFHITFVNNEFNHKRLIKSLGHDFLKGQPDFRFETI 313
Query: 54 GSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSI-----IVYDTLLYWAVV 108
+ PS+ + +E P +EL++ L++ + I+YD L+ +AV
Sbjct: 314 PDGLPPSDENATQSIGGLVEGCRKHCYGPLKELVENLNNSSEVPQVTSIIYDGLMGFAVD 493
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVK----------------YPGFEW 152
VA I FW A +L + P K PG +
Sbjct: 494 VAKDLGIAEQQFWTASACGLMGYLQFDELIKRNMLPYKDESYITDGSLDVHLDWTPGMKN 673
Query: 153 IH-----------------------------KAQYLLFSSIYELESQAIDVLKSKLPLPI 183
I K+ ++ +++ ELES+ +D L + P I
Sbjct: 674 IRMRDLPSFVRTTTLDEISFVGFGLEAQQCMKSSAIIINTVKELESEVLDALMAINP-NI 850
Query: 184 YTIGPTIPKFSLIKNDPKPSN----------TNHSYYIEWLDSQPIGSVLYIAQGSFFSV 233
Y IGP +F L N P+ N N I+WLD SV+YI GS +
Sbjct: 851 YNIGPL--QF-LANNFPEKENGFKSNGSSLWKNDLTCIKWLDQWEPSSVIYINYGSIAVM 1021
Query: 234 SSAQIDEIAAALCASNVRFLWIARSEASRLK-------EICGAHHMGLIMEWCDQLRVLS 286
S +E A L S + FLWI R + + K + G I+ WC Q VL+
Sbjct: 1022SEKHFNEFAWGLANSKLPFLWINRPDLVKGKCRPLSQEFLDEVKDRGYIITWCPQSEVLA 1201
Query: 287 HPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDT 346
HPS+G F +HCGWNS+ ES+ G P + P + +Q N + + W +G +K+DVKR
Sbjct: 1202HPSVGVFLTHCGWNSSLESISEGKPMIGWPFFAEQQTNCRYISTTWGIGMDIKDDVKR-- 1375
Query: 347 LVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMH 405
+++ +LV E + GE + RE+ + +K + + GGS++ DF + D H
Sbjct: 1376----EEVTELVKEM--IKGEKGKKKREKCVEWKKKVVEAAKPGGSSYNDFYRLLKDAYH 1534
>AW268009 weakly similar to PIR|T00583|T005 probable indole-3-acetate
beta-glucosyltransferase T27E13.11 - Arabidopsis
thaliana, partial (17%)
Length = 621
Score = 146 bits (368), Expect = 2e-35
Identities = 83/163 (50%), Positives = 101/163 (61%), Gaps = 7/163 (4%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P+P RG+INPMMNL KLLVSNN NI V+FVVT+EWL+ I SEPKPDNI S NV+
Sbjct: 71 VPYPSRGHINPMMNLSKLLVSNNPNILVTFVVTQEWLTLIDSEPKPDNIRVESIPNVV-- 244
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPP-SIIVYDTLLYWAVVVANRRNIPAAL 119
G +E VMT+MEAPFE L+D L PP + I+ D L+WA+ V NRRNIP A
Sbjct: 245 ----GDKFMDVVEAVMTEMEAPFERLIDRLVRPPVTFIICDCFLFWAIRVGNRRNIPVAA 412
Query: 120 FWPMPAS-----IFSVFLHQHIFEQNGHYPVKY-PGFEWIHKA 156
FW S F +FL + E NG Y + Y P W+ A
Sbjct: 413 FWTTSTSELWVQFFHIFLQRKNLE-NGEYYIDYIPSNSWVRLA 538
>TC82528 weakly similar to PIR|C86356|C86356 hypothetical protein AAF87257.1
[imported] - Arabidopsis thaliana, partial (35%)
Length = 1376
Score = 144 bits (363), Expect = 6e-35
Identities = 90/268 (33%), Positives = 143/268 (52%), Gaps = 17/268 (6%)
Frame = +1
Query: 154 HKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSYY--- 210
H+A ++F++ ELES ++ L S P +Y+IGP S ++ S +++ +
Sbjct: 496 HRASAIVFNTYNELESDVLNALHSMFP-SLYSIGPLPSLLSQTPHNHLESLSSNLWKEDT 672
Query: 211 --IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASRLKEICG 268
+EWL+S+ SV+Y+ GS ++ Q+ E A L S FLWI R + + G
Sbjct: 673 KCLEWLESKEPESVVYVNFGSITVMTPNQLLEFAWGLADSKKPFLWIIRPDL-----VIG 837
Query: 269 AHHM------------GLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLP 316
+ GLI WC Q +VL HPSIGGF +HCGWNST ES+ AGVP L P
Sbjct: 838 GSFILSSEFENEISDRGLITSWCPQEQVLIHPSIGGFLTHCGWNSTTESICAGVPMLCWP 1017
Query: 317 IYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSK 376
+ DQP N + + +W++G + DVKR D++ KLV+E GE + +R+++
Sbjct: 1018FFGDQPTNCRFICNEWEIGLEIDMDVKR------DEVEKLVNEL--TVGEKGKKMRQKAV 1173
Query: 377 KLQKICLNSIANGGSAHTDFNAFISDVM 404
+L+K + GG ++ + + I +V+
Sbjct: 1174ELKKKAEENTRPGGRSYMNLDKVIKEVL 1257
>TC82794 weakly similar to GP|3928543|dbj|BAA34687.1 UDP-glucose
glucosyltransferase {Arabidopsis thaliana}, partial
(59%)
Length = 1309
Score = 130 bits (326), Expect(2) = 1e-34
Identities = 101/370 (27%), Positives = 154/370 (41%), Gaps = 68/370 (18%)
Frame = +1
Query: 43 EPKPDNISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDH--------PP 94
E PD ++ G + ++I + + + PF ELL L P
Sbjct: 178 ETIPDGLTPIEGDGDVSQDII------SLSDSIRKNFYHPFCELLARLKDSSNDGHIPPV 339
Query: 95 SIIVYDTLLYWAVVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVKYPGF---- 150
S +V D L + + A +P+ LF A LH G P+K +
Sbjct: 340 SCLVSDIGLTFTIQAAEEHGLPSVLFSSASACSLLSALHFRTLIDKGVIPLKDESYLTNG 519
Query: 151 ------EWI-----------------------------------HKAQYLLFSSIYELES 169
+WI H+A ++F++ ELE+
Sbjct: 520 YLDTKVDWIPGLGNLRLKDLPDFIRTTDPNDIMIKFIIEAADRVHEANSIVFNTSDELEN 699
Query: 170 QAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTN--------HSYYIEWLDSQPIGS 221
I+ L K+P IY IGP S + P+ + + +EWL+S+ GS
Sbjct: 700 DVINALSIKIP-SIYAIGPLT---SFLNQSPQNNLASIGSNLWKEDMKCLEWLESKEQGS 867
Query: 222 VLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR-------SEASRLKEICGAHHMGL 274
V+Y+ GS ++ Q+ E A L S FLWI R S + G+
Sbjct: 868 VVYVNFGSITVMTPDQLLEFAWGLANSKKPFLWIIRPDLVIGGSVILSSDFVNETSDRGV 1047
Query: 275 IMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKV 334
I WC Q +VL+HPS+GGF +HCGWNST ES+ AGVP L P + +QP N + + +W++
Sbjct: 1048IASWCPQEKVLNHPSVGGFLTHCGWNSTMESICAGVPMLCWPFFAEQPTNCRYICNEWEI 1227
Query: 335 GCRVKEDVKR 344
G + +VK+
Sbjct: 1228GAEIDTNVKK 1257
Score = 34.3 bits (77), Expect(2) = 1e-34
Identities = 15/33 (45%), Positives = 24/33 (72%)
Frame = +3
Query: 2 PFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTE 34
P+P++G+INP++ L KLL + H++FV TE
Sbjct: 15 PYPLQGHINPLLKLAKLL--HLRGFHITFVNTE 107
>TC85817 weakly similar to PIR|B85014|B85014 probable flavonol
glucosyltransferase [imported] - Arabidopsis thaliana,
partial (83%)
Length = 1736
Score = 143 bits (360), Expect = 1e-34
Identities = 131/467 (28%), Positives = 208/467 (44%), Gaps = 75/467 (16%)
Frame = +3
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEW------LSFISSEPKPDNISF--- 51
+P P G++ PM+ K ++ N N+ ++F + E + + S PK + +F
Sbjct: 171 LPSPGMGHLIPMIEFAKRIIILNQNLQITFFIPTEGPPSKAQKTVLQSLPKFISHTFLPP 350
Query: 52 --------RSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLL 103
SG I S + R P+ ++ T E H + +V D
Sbjct: 351 VSFSDLPPNSGIETIISLTVL-RSLPSLRQNFNTLSET----------HTITAVVVDLFG 497
Query: 104 YWAVVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHY-------PVKYPGFEWIH-- 154
A VA N+P +F+P A S+FL+ ++ H PVK PG IH
Sbjct: 498 TDAFDVAREFNVPKYVFYPSTAMALSLFLYLPRLDEEVHCEFRELTEPVKIPGCIPIHGK 677
Query: 155 -------------------------KAQYLLFSSIYELESQAI-DVLKSKLPLP-IYTIG 187
+A L+ +S ELE I ++LK + P Y +G
Sbjct: 678 YLLDPLQDRKNDAYQSVFRNAKRYREADGLIENSFLELEPGPIKELLKEEPGKPKFYPVG 857
Query: 188 PTIPKFSLIKNDPKPSNTN-HSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALC 246
P L+K + + +S ++WLD+QP GSVL+++ GS ++SS QI E+A L
Sbjct: 858 P------LVKREVEVGQIGPNSESLKWLDNQPHGSVLFVSFGSGGTLSSKQIVELALGLE 1019
Query: 247 ASNVRFLWIARSEASRLKEIC--------------------GAHHMGLIME-WCDQLRVL 285
S RFLW+ RS ++ GL++ W Q +VL
Sbjct: 1020MSEQRFLWVVRSPNDKVANASYFSAETDSDPFDFLPNGFLERTKGRGLVVSSWAPQPQVL 1199
Query: 286 SHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRD 345
+H S GGF +HCGWNS ES+V GVP + P+Y +Q N+ M+ ED KVG R +V +
Sbjct: 1200AHGSTGGFLTHCGWNSVLESVVNGVPLVVWPLYAEQKMNAVMLTEDVKVGLR--PNVGEN 1373
Query: 346 TLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSA 392
LV++ +I +V M +GE + +R + K L++ ++ G++
Sbjct: 1374GLVERLEIASVVKCLM--EGEEGKKLRYQMKDLKEAASKTLGENGTS 1508
>TC90534 weakly similar to GP|19911197|dbj|BAB86925. glucosyltransferase-7
{Vigna angularis}, partial (71%)
Length = 808
Score = 137 bits (344), Expect = 1e-32
Identities = 78/229 (34%), Positives = 127/229 (55%), Gaps = 7/229 (3%)
Frame = +3
Query: 185 TIGPTIPKFSLIKN-----DPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQID 239
T+GP +P L K D + IEWL+++P S +Y++ GS S++ QI+
Sbjct: 36 TVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIE 215
Query: 240 EIAAALCASNVRFLWIAR-SEASRL-KEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHC 297
E+A L FLW+ + SE ++L K+ GLI+ WC QL VL+H +IG F +HC
Sbjct: 216 EVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSENGLIVAWCPQLEVLAHEAIGCFVTHC 395
Query: 298 GWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLV 357
GWNST E+L GVP + +P+Y DQ ++K +V+ WKVG R D K+ +V+KD + +
Sbjct: 396 GWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQ--IVRKDPLKDCI 569
Query: 358 HEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMHL 406
E M + E ++I + + + ++ GS+H + F++ + +
Sbjct: 570 CEIMSM-SEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSLFQV 713
>TC78695 similar to GP|4115563|dbj|BAA36423.1 UDP-glucose:anthocyanin
5-O-glucosyltransferase {Verbena x hybrida}, partial
(49%)
Length = 1741
Score = 135 bits (339), Expect = 4e-32
Identities = 72/207 (34%), Positives = 120/207 (57%), Gaps = 13/207 (6%)
Frame = +1
Query: 210 YIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR------- 262
YI+WLDS+ SV+Y++ G+ +S Q++EIA AL S FLW+ R + +
Sbjct: 874 YIQWLDSKDEKSVVYVSFGTLAVLSKRQMEEIARALLDSGFSFLWVIRDKKLQQQKEEEV 1053
Query: 263 ------LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLP 316
+E + G I++WC Q+ VLSH S+G F +HCGWNST ESL +GVP + P
Sbjct: 1054 DDDELSCREELENNMNGKIVKWCSQVEVLSHRSLGCFMTHCGWNSTLESLGSGVPMVAFP 1233
Query: 317 IYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSK 376
+ DQ N+K++ + WK G R++ D + +VK ++I K + M GE ++R +K
Sbjct: 1234 QWTDQTTNAKLIEDVWKTGLRMEHD--EEGMVKVEEIRKCLEVVMG-KGEKGEELRRNAK 1404
Query: 377 KLQKICLNSIANGGSAHTDFNAFISDV 403
K + + ++ GGS++ + ++++D+
Sbjct: 1405 KWKDLARAAVKEGGSSNRNLRSYLNDI 1485
Score = 57.8 bits (138), Expect = 8e-09
Identities = 59/251 (23%), Positives = 114/251 (44%), Gaps = 20/251 (7%)
Frame = +2
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS-- 60
+P++G+INP + K L+S + V+F T S + ++P +SF + S+
Sbjct: 101 YPLQGHINPALQFTKRLISLGAK--VTFATTIHLYSRLINKPTIPGLSFATFSDGYDDGQ 274
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLL----DHPPSIIVYDTLLYWAVVVANRRNIP 116
+ D ++M + T+ + F + L +HP + ++Y +L WA VA+ ++P
Sbjct: 275 KSFGDEDIVSYMSE-FTRRGSEFLTNIILSSKQENHPFTCLIYTLILSWAPKVAHELHLP 451
Query: 117 AALFWPMPASIFSVFLHQHIFEQNGHY----------PVKYPGFEWIHKAQ----YLLFS 162
+ L W A++F +F + F ++G Y + PG + K++ +LL S
Sbjct: 452 STLLWIQAATVFDIF--YYYFHEHGDYITNKSKDETCLISLPGLSFSLKSRDLPSFLLAS 625
Query: 163 SIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSYYIEWLDSQPIGSV 222
+ Y A+ LK ++ L I P + L+ NT + ++ L+ +G +
Sbjct: 626 NTYTF---ALPSLKEQIQLLNEEINPRV----LV-------NTVEEFELDALNKVDVGKI 763
Query: 223 LYIAQGSFFSV 233
I G+ S+
Sbjct: 764 KMIPIGTVDSI 796
>TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyltransferase
HRA25 {Phaseolus vulgaris}, partial (47%)
Length = 1332
Score = 134 bits (336), Expect = 8e-32
Identities = 124/466 (26%), Positives = 207/466 (43%), Gaps = 62/466 (13%)
Frame = +1
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTE--------------EWLSFISS---- 42
+P+P+ G+INP+M LC +L + I +F+ TE E ++F++
Sbjct: 28 IPYPIPGHINPLMQLCHVLAKHGCKI--TFLNTEFSHKRTNNNNEQSQETINFVTLPDGL 201
Query: 43 EPKPDNISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTL 102
EP+ D RS + + R+ P + ++ ++ A +E + I+
Sbjct: 202 EPEDD----RSDQKKVLFSI--KRNMPPLLPKLIEEVNALDDE------NKICCIIVTFN 345
Query: 103 LYWAVVVANRRNIPAALFWPMPASIFSV------FLHQHIFEQNGHYP------------ 144
+ WA+ V + I L W A+ + + + + G Y
Sbjct: 346 MGWALEVGHNLGIKGVLLWTGSATSLAFCYSIPKLIDDGVIDSAGIYTKDQEIQLSPNMP 525
Query: 145 ------VKYPGFEWI---HKAQYL---------LFSSIYELESQAIDVLKSKLPLPIYTI 186
V + F+ I H AQ + L ++ Y+LE + LP I
Sbjct: 526 KMDTKNVPWRTFDKIIFDHLAQQMQTMKLGHWWLCNTTYDLEHATFSISPKFLP-----I 690
Query: 187 GPTIPKFSLIKNDPKPSN--TNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAA 244
GP L++ND S+ ++WLD QP SV+Y++ GS + Q +E+A
Sbjct: 691 GP------LMENDSNKSSFWQEDMTSLDWLDKQPSQSVVYVSFGSLAVMDQNQFNELALG 852
Query: 245 LCASNVRFLWIAR-SEASRLK-----EICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCG 298
L + FLW+ R S +++ E G G I+ W Q ++L+HP+I F SHCG
Sbjct: 853 LDLLDKPFLWVVRPSNDNKVNYAYPDEFLGTK--GKIVSWVPQKKILNHPAIACFISHCG 1026
Query: 299 WNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVH 358
WNST E + +G+PFL P DQ N + + WKVG + +D + +V K++I K V
Sbjct: 1027WNSTIEGVYSGIPFLCWPFATDQFTNKSYICDVWKVGFELDKD--ENGIVLKEEIKKKVE 1200
Query: 359 EFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
+ + +DI+ERS KL+++ L +I G + + FI+ M
Sbjct: 1201QLLQ-----DQDIKERSLKLKELTLENIVEDGKSSKNLQNFINWAM 1323
>TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase-8 {Vigna
angularis}, partial (53%)
Length = 1787
Score = 131 bits (330), Expect = 4e-31
Identities = 123/470 (26%), Positives = 200/470 (42%), Gaps = 68/470 (14%)
Frame = +1
Query: 2 PFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPSE 61
PF G+I P ++L ++ S + V+ V T + IS I+ ++ P E
Sbjct: 73 PFLANGHIIPCVDLARVFSSRG--LKVTIVTTHLNVPLISRTIGKAKINIKTIKFPSPEE 246
Query: 62 LI----CGRDHPAFMEDVMTK-------MEAPFEELLDLLDHPPSIIVYDTLLYWAVVVA 110
C A D K + P E +L+ P +V D W+ A
Sbjct: 247 TGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLE--QEKPDCLVADMFFPWSTDSA 420
Query: 111 NRRNIPAALFWPM---PASIFSVFLHQHIFEQNGHYPVKY-----PGFEWIHKAQY---- 158
+ NIP +F + P + + ++ Y + PG + K Q
Sbjct: 421 AKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLPGEITLTKMQLPQLP 600
Query: 159 ----------------------LLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLI 196
++ ++ YELE D +++L + +GP SL
Sbjct: 601 QHDKVFTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNELGRKAWHLGPV----SLC 768
Query: 197 KNDP-------KPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASN 249
D + ++ + ++WL S+ SV+Y+ GS S AQ+ EIA L AS
Sbjct: 769 NRDTEEKACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVFSDAQLKEIAMGLEASE 948
Query: 250 VRFLWI----ARSEASRLK--------EICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHC 297
V F+W+ A+SE L+ I G+ +I W Q+ +L H S+GGF +HC
Sbjct: 949 VPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQVMILDHESVGGFVTHC 1128
Query: 298 GWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKED----VKRDTLVKKDKI 353
GWNST E + AG+P +T P+Y +Q +N+K + + K+G V + VKKD I
Sbjct: 1129GWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQTWIGMGGGEPVKKDVI 1308
Query: 354 VKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
K V M G+ ++R R+K+ K+ ++ GGS++ DF+ I D+
Sbjct: 1309EKAVRRIM--VGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNLIEDL 1452
>AW776615 weakly similar to GP|14532546|gb At1g22370/T16E15_3 {Arabidopsis
thaliana}, partial (48%)
Length = 708
Score = 128 bits (321), Expect = 5e-30
Identities = 69/191 (36%), Positives = 112/191 (58%), Gaps = 7/191 (3%)
Frame = -2
Query: 221 SVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSE----ASRLKEICGAHHM---G 273
SV+Y+ GS ++ Q+ E A L S FLWI R + S + A+ + G
Sbjct: 701 SVVYVNFGSITVMTPEQLQEFAWGLANSKKPFLWITRPDLVIGGSVILSSDFANEISDRG 522
Query: 274 LIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWK 333
LI WC Q +VL+HPSIGGF +HCGWNST ES+ AGVP L P + DQP + + + +WK
Sbjct: 521 LIASWCPQEKVLNHPSIGGFLTHCGWNSTTESICAGVPMLCWPFFADQPTDCRFICNEWK 342
Query: 334 VGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAH 393
+G + DT VK++++ KL++E + G+ +++RE++ +L+K + GG ++
Sbjct: 341 IGMEI------DTNVKREEVAKLINEL--IAGDEGKNMREKAMELKKAAEENTRPGGCSY 186
Query: 394 TDFNAFISDVM 404
+F+ I +++
Sbjct: 185 MNFDKVIKEML 153
>TC89558 similar to GP|14349253|dbj|BAB60721. glucosyltransferase {Nicotiana
tabacum}, partial (35%)
Length = 873
Score = 124 bits (310), Expect = 9e-29
Identities = 73/232 (31%), Positives = 117/232 (49%), Gaps = 25/232 (10%)
Frame = +2
Query: 199 DPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARS 258
+PK + S I WLD QP+ SV+++ GS Q+ EIA A+ S F+W R
Sbjct: 53 EPKTKDVVGSDIINWLDDQPLSSVVFLCLGSRGCFDEDQVKEIAHAIENSGAHFVWSLRK 232
Query: 259 EASR----------LKEICGA---------HHMGLIMEWCDQLRVLSHPSIGGFWSHCGW 299
A + L ++C +G ++ W Q ++L+HP+IGGF SHCGW
Sbjct: 233 PAPKGAMAAPSDYTLSDLCSVLPEGFLDRTEEIGRVIGWAPQAQILAHPAIGGFVSHCGW 412
Query: 300 NSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVK------RDTLVKKDKI 353
NST ES+ GVP T P++ +Q N+ +V + K+ + D + + L+ DKI
Sbjct: 413 NSTLESIYFGVPIATWPLFAEQQVNAFELVCELKISVEIALDYRVEFNSGPNYLLTADKI 592
Query: 354 VKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMH 405
K + +D DGE + ++E S+K +K ++ GGS+ + I +M+
Sbjct: 593 EKGIRSVLDKDGEFRKKMKEMSEKSKK----TLLEGGSSSIYLSRLIDYIMN 736
>TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransferase {Vigna
angularis}, partial (87%)
Length = 1704
Score = 123 bits (308), Expect = 1e-28
Identities = 84/266 (31%), Positives = 134/266 (49%), Gaps = 25/266 (9%)
Frame = +3
Query: 160 LFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLI---KNDPKPSNTNHSYYIEWLDS 216
L +S Y+LE D +++KL + +GP + K K + + WL+S
Sbjct: 678 LINSFYDLEPAYADYVRNKLGKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNS 857
Query: 217 QPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWI-------ARSEAS-------- 261
+ SVLYI+ GS V Q+ EIA L AS+ F+W+ +++E
Sbjct: 858 KKPNSVLYISFGSVARVPMKQLKEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKFER 1037
Query: 262 RLKEICGAHHMGLIME-WCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYID 320
R+KE+ GLI W QL +L H ++GGF +HCGWNST E + AGVP T P+ +
Sbjct: 1038RMKEM----DKGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAE 1205
Query: 321 QPFNSKMMVEDWKVGCRVKE------DVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRER 374
Q N K++ + ++G +V D +R LV ++K+ V + M + E T ++R R
Sbjct: 1206QFINEKLITDVLRIGVQVGSREWGSWDEERKELVGREKVELAVKKLM-AESEDTEEMRRR 1382
Query: 375 SKKLQKICLNSIANGGSAHTDFNAFI 400
K + + ++ GGS++ D +A I
Sbjct: 1383VKSIFENAKRAVEEGGSSYDDIHALI 1460
>BQ123034 weakly similar to GP|14334982|gb putative glucosyltransferase
{Arabidopsis thaliana}, partial (24%)
Length = 765
Score = 123 bits (308), Expect = 1e-28
Identities = 73/220 (33%), Positives = 122/220 (55%), Gaps = 20/220 (9%)
Frame = +2
Query: 207 HSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEAS----- 261
H+ + WL+S+ SVLY++ GSF + AQ+ EI L S F+W+ + + +
Sbjct: 68 HTELLNWLNSKENESVLYVSFGSFTRLPYAQLVEIVHGLENSGHNFIWVIKRDDTDEDGE 247
Query: 262 --------RLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFL 313
R+KE + +I +W QL +L HP+ GG +HCGWNST ESL AG+P +
Sbjct: 248 GFLQEFEERIKESSKGY---IIWDWAPQLLILDHPATGGIVTHCGWNSTLESLNAGLPMI 418
Query: 314 TLPIYIDQPFNSKMMVEDWKVGCRV--KE-----DVKRDTLVKKDKIVKLVHEFMDLDGE 366
T PI+ +Q +N K++V+ K+G V KE D+ + +V++++I K V M G+
Sbjct: 419 TWPIFAEQFYNEKLLVDVLKIGVPVGAKENKLWLDISVEKVVRREEIEKTVKILMG-SGQ 595
Query: 367 LTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMHL 406
++++R R+KKL + +I GG ++ + I ++ L
Sbjct: 596 ESKEMRMRAKKLSEAAKRTIEEGGDSYNNLIQLIDELKSL 715
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,237,129
Number of Sequences: 36976
Number of extensions: 245949
Number of successful extensions: 1439
Number of sequences better than 10.0: 156
Number of HSP's better than 10.0 without gapping: 1340
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1365
length of query: 406
length of database: 9,014,727
effective HSP length: 98
effective length of query: 308
effective length of database: 5,391,079
effective search space: 1660452332
effective search space used: 1660452332
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141923.7


