

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.5 - phase: 0 /pseudo
(648 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
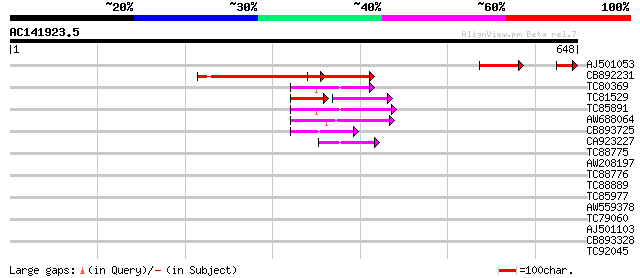
Score E
Sequences producing significant alignments: (bits) Value
AJ501053 similar to GP|17064832|gb putative VP1/ABI3 family regu... 111 3e-33
CB892231 similar to GP|7270105|emb predicted protein {Arabidopsi... 134 1e-31
TC80369 similar to PIR|A86386|A86386 probable DNA-binding protei... 74 1e-13
TC81529 similar to GP|22335711|dbj|BAC10553. ABA insensitive 3 {... 44 3e-11
TC85891 similar to PIR|A86386|A86386 probable DNA-binding protei... 65 1e-10
AW688064 homologue to GP|13272405|gb unknown protein {Arabidopsi... 57 2e-08
CB893725 weakly similar to GP|11932104|emb VP1/ABI3-like protein... 52 5e-07
CA923227 similar to GP|20152530|emb putative auxin response fact... 42 6e-04
TC88775 homologue to PIR|F86427|F86427 auxin response factor 6 (... 41 0.002
AW208197 similar to PIR|F84776|F847 probable RAV2-like DNA bindi... 40 0.002
TC88776 homologue to PIR|F86427|F86427 auxin response factor 6 (... 36 0.053
TC88889 similar to GP|20340241|gb|AAM19707.1 putative RING zinc ... 31 1.3
TC85977 similar to GP|10176918|dbj|BAB10162. auxin response fact... 30 2.2
AW559378 similar to GP|9955527|emb potassium transport protein-l... 30 3.8
TC79060 similar to GP|1633130|pdb|1SCH|A Chain A Peanut Peroxid... 30 3.8
AJ501103 29 4.9
CB893328 similar to PIR|D71431|D714 hypothetical protein - Arabi... 29 6.5
TC92045 28 8.4
>AJ501053 similar to GP|17064832|gb putative VP1/ABI3 family regulatory
protein {Arabidopsis thaliana}, partial (13%)
Length = 682
Score = 111 bits (277), Expect(2) = 3e-33
Identities = 51/51 (100%), Positives = 51/51 (100%)
Frame = +3
Query: 537 ELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGKIKTNASPSG 587
ELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGKIKTNASPSG
Sbjct: 3 ELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGKIKTNASPSG 155
Score = 49.7 bits (117), Expect(2) = 3e-33
Identities = 23/24 (95%), Positives = 24/24 (99%)
Frame = +2
Query: 625 QVFRFCIRAARSKGTRKSSKNQKR 648
+VFRFCIRAARSKGTRKSSKNQKR
Sbjct: 152 RVFRFCIRAARSKGTRKSSKNQKR 223
>CB892231 similar to GP|7270105|emb predicted protein {Arabidopsis thaliana},
partial (21%)
Length = 815
Score = 134 bits (336), Expect = 1e-31
Identities = 73/147 (49%), Positives = 96/147 (64%), Gaps = 1/147 (0%)
Frame = +2
Query: 215 SLKMALGTSSRNSVLPLATEIGEGKLEGKASSHFQQGQTSQSILAQLSKTGIAMNLETNK 274
+L M L N P + + + + K S S+ +L + + ++ LE N
Sbjct: 176 NLSMTLAAPLPN---PFHNVLVDEREQSKMSPPLLLASRSRHLLPKPPRPALSPGLEGNT 346
Query: 275 GMISHPP-RRPPADVKGKNQLLSRYWPRITDQELEKLSGDLKSTVVPLFEKVLSPSDAGR 333
GM+S RPPA+ +G+NQLL RYWPRITDQEL+++SGD ST+VPLFEK+LS SDAGR
Sbjct: 347 GMVSQIRIARPPAEGRGRNQLLPRYWPRITDQELQQISGDSNSTIVPLFEKMLSASDAGR 526
Query: 334 IGRLVLPKACAEAFLPRILQSEGVPLQ 360
IGRLVLPKACAEA+ P I Q EG+PL+
Sbjct: 527 IGRLVLPKACAEAYFPPISQPEGLPLR 607
Score = 105 bits (263), Expect = 4e-23
Identities = 48/77 (62%), Positives = 57/77 (73%)
Frame = +1
Query: 341 KACAEAFLPRILQSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNA 400
K+ + P L + G +QD+ G EW FQFRFWPNNNSRMYVLEGVTPCIQS+QL A
Sbjct: 547 KSMC*SIFPSYLST*GPSFAYQDVKGKEWMFQFRFWPNNNSRMYVLEGVTPCIQSMQLQA 726
Query: 401 GDTVTFSRIDPGEKFLF 417
GDTVTFSR+DP K ++
Sbjct: 727 GDTVTFSRMDPEGKLIW 777
>TC80369 similar to PIR|A86386|A86386 probable DNA-binding protein RAV2
[imported] - Arabidopsis thaliana, partial (30%)
Length = 821
Score = 74.3 bits (181), Expect = 1e-13
Identities = 45/108 (41%), Positives = 61/108 (55%), Gaps = 11/108 (10%)
Frame = +3
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLP----------RILQSEGVPLQFQDIMGNEWT 370
LFEKV++PSD G++ RLV+PK AE P +GV L F+DI G W
Sbjct: 15 LFEKVVTPSDVGKLNRLVIPKQHAEKHFPLQKADCVQGSASAAGKGVLLNFEDIGGKVWR 194
Query: 371 FQFRFWPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSRIDPGEKFLF 417
F++ +W N+S+ YVL +G + ++ L AGDTV F R EK LF
Sbjct: 195 FRYSYW--NSSQSYVLTKGWSRFVKEKNLKAGDTVCFQRSTGPEKQLF 332
>TC81529 similar to GP|22335711|dbj|BAC10553. ABA insensitive 3 {Pisum
sativum}, partial (37%)
Length = 1075
Score = 43.9 bits (102), Expect(2) = 3e-11
Identities = 20/44 (45%), Positives = 28/44 (63%)
Frame = +1
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDI 364
L +KVL SD G +GR+VLPK AE LP + +G+ + +DI
Sbjct: 550 LLQKVLKQSDVGSLGRIVLPKKEAETHLPELEARDGISITMEDI 681
Score = 42.4 bits (98), Expect(2) = 3e-11
Identities = 23/70 (32%), Positives = 35/70 (49%), Gaps = 1/70 (1%)
Frame = +2
Query: 369 WTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFGFRRSLTSIV 427
W ++R+WPNN SRMY+LE +++ L GD V +S + G+ + G T
Sbjct: 698 WNMRYRYWPNNKSRMYLLENTGDFVKANGLQEGDFIVIYSDVKCGKFMIRGR*SEATREQ 877
Query: 428 TQDASTSSHS 437
Q A + S
Sbjct: 878 NQKAKKAGKS 907
>TC85891 similar to PIR|A86386|A86386 probable DNA-binding protein RAV2
[imported] - Arabidopsis thaliana, partial (57%)
Length = 2083
Score = 64.7 bits (156), Expect = 1e-10
Identities = 43/140 (30%), Positives = 73/140 (51%), Gaps = 18/140 (12%)
Frame = +2
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLP---------------RILQSEGVPLQFQDIM 365
LFEK ++PSD G++ RLV+PK AE P ++G+ L F+D+
Sbjct: 1160 LFEKTVTPSDVGKLNRLVIPKQHAEKHFPLNAVAVAVACDGVSTAAAAAKGLLLNFEDVG 1339
Query: 366 GNEWTFQFRFWPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSRIDPGEKFLF--GFRRS 422
G W F++ +W N+S+ YVL +G + ++ L AGD V F R ++ L+ RS
Sbjct: 1340 GKVWRFRYSYW--NSSQSYVLTKGWSRFVKEKNLRAGDAVRFFRSTGPDRQLYIDCKARS 1513
Query: 423 LTSIVTQDASTSSHSNGILI 442
+ + Q + ++++ G+ I
Sbjct: 1514 IGVVGGQVDNNNNNTGGLFI 1573
>AW688064 homologue to GP|13272405|gb unknown protein {Arabidopsis thaliana},
partial (15%)
Length = 576
Score = 57.4 bits (137), Expect = 2e-08
Identities = 39/121 (32%), Positives = 59/121 (48%), Gaps = 3/121 (2%)
Frame = +1
Query: 322 FEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQ---FQDIMGNEWTFQFRFWPN 378
F K L+ SDA G +P+ CAE PR+ S P+Q +D+ G W F+ +
Sbjct: 97 FAKTLTQSDANNGGGFSVPRYCAETIFPRLDYSAEPPVQTVIAKDVHGEVWKFR-HIYRG 273
Query: 379 NNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSN 438
R + G + + +L AGD++ F R + GE F+ G RR+ IV + S S+
Sbjct: 274 TPRRHLLTTGWSSFVNQKKLVAGDSIVFLRAESGELFV-GIRRAKRGIVNGLETPSGWSS 450
Query: 439 G 439
G
Sbjct: 451 G 453
>CB893725 weakly similar to GP|11932104|emb VP1/ABI3-like protein
{Chamaecyparis nootkatensis}, partial (3%)
Length = 793
Score = 52.4 bits (124), Expect = 5e-07
Identities = 29/79 (36%), Positives = 42/79 (52%), Gaps = 1/79 (1%)
Frame = +3
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIM-GNEWTFQFRFWPNN 379
+ K L SD GR+VLPK E LP + EG+ + F+DI G EW + ++W N
Sbjct: 501 ILTKKLRKSDVKNYGRIVLPKREVEEKLPAPSR-EGIEVIFKDIYSGREWKLKLKYWING 677
Query: 380 NSRMYVLEGVTPCIQSLQL 398
+RMY+LE + +L
Sbjct: 678 TTRMYILENTGDLVNHYKL 734
>CA923227 similar to GP|20152530|emb putative auxin response factor 32
{Arabidopsis thaliana}, partial (25%)
Length = 238
Score = 42.4 bits (98), Expect = 6e-04
Identities = 26/73 (35%), Positives = 43/73 (58%), Gaps = 3/73 (4%)
Frame = -3
Query: 353 QSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFS--RI 409
+ +G+ L F+D G W F++ +W N+S+ YVL +G + ++ +L+AGD V F R+
Sbjct: 221 ECKGLLLSFEDESGKCWRFRYFYW--NSSQSYVLTKGWSRYVKDKRLDAGDVVLFERHRV 48
Query: 410 DPGEKFLFGFRRS 422
D F+ RRS
Sbjct: 47 DSQRLFINWRRRS 9
>TC88775 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (23%)
Length = 678
Score = 40.8 bits (94), Expect = 0.002
Identities = 32/104 (30%), Positives = 49/104 (46%), Gaps = 3/104 (2%)
Frame = +3
Query: 322 FEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQ---FQDIMGNEWTFQFRFWPN 378
F K L+ SD G +P+ AE P + S+ P Q +D+ GNEW F+ F
Sbjct: 24 FCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIF-RG 200
Query: 379 NNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRRS 422
R + G + + + +L AGD+V F + + L G RR+
Sbjct: 201 QPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQ-LLLGIRRA 329
>AW208197 similar to PIR|F84776|F847 probable RAV2-like DNA binding protein
[imported] - Arabidopsis thaliana, partial (21%)
Length = 415
Score = 40.4 bits (93), Expect = 0.002
Identities = 24/55 (43%), Positives = 32/55 (57%), Gaps = 7/55 (12%)
Frame = +3
Query: 302 ITDQELEKLSGDLKSTVVP-------LFEKVLSPSDAGRIGRLVLPKACAEAFLP 349
I D+EL D +TVV +FEK L+PSD G++ RLV+PK AE + P
Sbjct: 255 IEDEEL-----DTTTTVVEPEDEKESMFEKPLTPSDVGKLNRLVIPKQYAEKYFP 404
>TC88776 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (22%)
Length = 1278
Score = 35.8 bits (81), Expect = 0.053
Identities = 25/75 (33%), Positives = 36/75 (47%), Gaps = 3/75 (4%)
Frame = +1
Query: 301 RITDQELEKLSGDLKSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQ 360
R+T Q+ +L G + F K L+ SD G +P+ AE P + S+ P Q
Sbjct: 841 RLTFQQSWELQGKQPTNY---FCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQ 1011
Query: 361 ---FQDIMGNEWTFQ 372
+D+ GNEW FQ
Sbjct: 1012ELIARDLHGNEWKFQ 1056
>TC88889 similar to GP|20340241|gb|AAM19707.1 putative RING zinc finger
protein-like protein {Thellungiella halophila}, partial
(46%)
Length = 911
Score = 31.2 bits (69), Expect = 1.3
Identities = 17/65 (26%), Positives = 25/65 (38%)
Frame = +1
Query: 30 FAKLCNKCGLAYENSLFCDKFHRHETGWRKCSNCSKPIHSGCIVSKSSFEYLDFGGITCV 89
F +CN C + C E+ C +C H C+ ++LD+ ITC
Sbjct: 457 FDSVCNSCKEPEHDCSVCLTQFEPESEINYCISCGHVFHKVCLE-----KWLDYWNITCP 621
Query: 90 TCVKP 94
C P
Sbjct: 622 LCRSP 636
>TC85977 similar to GP|10176918|dbj|BAB10162. auxin response factor-like
protein {Arabidopsis thaliana}, partial (65%)
Length = 3162
Score = 30.4 bits (67), Expect = 2.2
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 3/57 (5%)
Frame = +3
Query: 322 FEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQ---FQDIMGNEWTFQFRF 375
F K L+ SD G + K A+ LP + S+ P Q +D+ GNEW F+ F
Sbjct: 615 FCKTLTASDTSTHGGFSVLKRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIF 785
>AW559378 similar to GP|9955527|emb potassium transport protein-like
{Arabidopsis thaliana}, partial (15%)
Length = 610
Score = 29.6 bits (65), Expect = 3.8
Identities = 11/33 (33%), Positives = 21/33 (63%)
Frame = -3
Query: 190 SHLTAFTTLENNRPSWETKSIDETLSLKMALGT 222
+HL + + +N P W + S+ E LSL+ ++G+
Sbjct: 149 THLDSVESKDNALPGWSSSSVIELLSLRFSIGS 51
>TC79060 similar to GP|1633130|pdb|1SCH|A Chain A Peanut Peroxidase,
partial (93%)
Length = 1412
Score = 29.6 bits (65), Expect = 3.8
Identities = 20/79 (25%), Positives = 30/79 (37%)
Frame = +1
Query: 63 CSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNTENHNRFSQTTKNNASDQYGEH 122
C + GCI + SSF L + C++ VK H F TK Q+ E
Sbjct: 253 CIERSSFGCIFASSSFSRLLCSRV*CISIVK---------GHTNFQGRTKCQTQCQFSEG 405
Query: 123 IDGRLLVEQAGKGNLMQLC 141
+ + + + L Q C
Sbjct: 406 LRNHRKCKSSIRATLSQCC 462
>AJ501103
Length = 576
Score = 29.3 bits (64), Expect = 4.9
Identities = 16/52 (30%), Positives = 29/52 (55%)
Frame = -2
Query: 504 GPLQRLVSVSEKKRSRNIGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSV 555
GP R + V +KR+ +G + ++IH +D + RL A++ C PP++
Sbjct: 155 GPFVRGLCVKGRKRAMGLG*RFDFVYIHFDDVCKERL-GRGAKKNRCTPPAI 3
>CB893328 similar to PIR|D71431|D714 hypothetical protein - Arabidopsis
thaliana, partial (36%)
Length = 892
Score = 28.9 bits (63), Expect = 6.5
Identities = 13/34 (38%), Positives = 22/34 (64%)
Frame = +3
Query: 250 QGQTSQSILAQLSKTGIAMNLETNKGMISHPPRR 283
QGQ + ++ Q+S G ++NLE+ + +PPRR
Sbjct: 645 QGQETLNLGTQVSHEGSSLNLESLAYKLDNPPRR 746
>TC92045
Length = 873
Score = 28.5 bits (62), Expect = 8.4
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = -3
Query: 92 VKPSQLCLNTENHNRFSQTTKNNAS 116
+ P + TENH +FS+ TKN +S
Sbjct: 700 LSPKSVRSKTENHQKFSKNTKNKSS 626
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,079,026
Number of Sequences: 36976
Number of extensions: 356625
Number of successful extensions: 1694
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1678
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1691
length of query: 648
length of database: 9,014,727
effective HSP length: 102
effective length of query: 546
effective length of database: 5,243,175
effective search space: 2862773550
effective search space used: 2862773550
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC141923.5


