

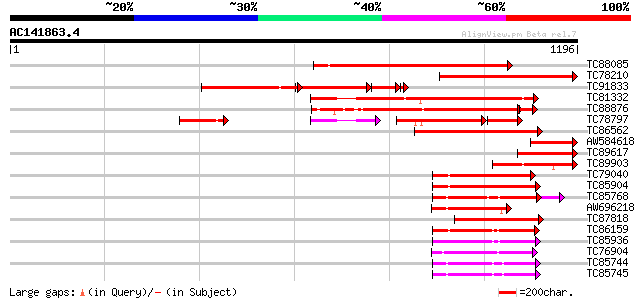
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141863.4 + phase: 0 /partial
(1196 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88085 similar to GP|20259341|gb|AAM13995.1 unknown protein {Ar... 634 0.0
TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Ar... 503 e-142
TC91833 similar to GP|20259341|gb|AAM13995.1 unknown protein {Ar... 390 e-108
TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 379 e-105
TC88876 similar to PIR|G96537|G96537 hypothetical protein F2J10.... 348 e-103
TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 206 9e-88
TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regu... 286 3e-77
AW584618 similar to GP|22831135|d putative MSP1(mitochondrial so... 199 4e-51
TC89617 similar to GP|22831135|dbj|BAC15996. putative MSP1(mitoc... 190 3e-48
TC89903 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 187 3e-47
TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase ... 182 7e-46
TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [i... 180 3e-45
TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle pr... 177 3e-44
AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33... 166 4e-41
TC87818 similar to GP|20197082|gb|AAC26698.2 putative katanin {A... 154 2e-37
TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AA... 150 2e-36
TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidops... 147 2e-35
TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein... 145 7e-35
TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15... 145 1e-34
TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome re... 144 2e-34
>TC88085 similar to GP|20259341|gb|AAM13995.1 unknown protein {Arabidopsis
thaliana}, partial (29%)
Length = 1261
Score = 634 bits (1634), Expect = 0.0
Identities = 317/421 (75%), Positives = 359/421 (84%), Gaps = 1/421 (0%)
Frame = +1
Query: 641 LFEVVTSESRDSPFILFMKEAEKSIVGNGDPYSFKSKLEKLPDNVVVIGSHTHSDSRKEK 700
+FE+ +S S+ +L +K+ EK + GN + KSK LP NVVVIGSH H D+RKEK
Sbjct: 4 IFEIASSLSKSGALVLLIKDIEKGVAGNSEV--LKSKFASLPQNVVVIGSHIHPDNRKEK 177
Query: 701 SHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDE 760
+ G LLFTKFG NQTALLDLAFPD+F RLHDR KE PK K L + FPNKVTI +PQDE
Sbjct: 178 TQPGSLLFTKFGGNQTALLDLAFPDNFTRLHDRSKETPKVMKQLNRFFPNKVTIQLPQDE 357
Query: 761 ALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKILG 820
ALL+ WKQ L+RDVET+K + N+ +R VL++ G++ LE+L +KD TLT EN EKI+G
Sbjct: 358 ALLSDWKQHLERDVETMKAQSNVVSIRLVLNKFGLDCPELETLSIKDQTLTTENVEKIIG 537
Query: 821 WALSHHLMQNPEADAD-AKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKR 879
WA+S+H M + EA + +K V+S+ESIQYG I Q IQNE+KS+KKSLKDVVTENEFEK+
Sbjct: 538 WAISYHFMHSSEASTEESKPVISAESIQYGFNILQGIQNENKSVKKSLKDVVTENEFEKK 717
Query: 880 LLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPP 939
LLGDVIPP DIGV+F+DIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPP
Sbjct: 718 LLGDVIPPTDIGVSFNDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPP 897
Query: 940 GTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVD 999
GTGKTMLAKAVAT+AGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVD
Sbjct: 898 GTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVD 1077
Query: 1000 SMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRR 1059
SMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD ERV+VLAATNRP+DLDEAVIRRLPRR
Sbjct: 1078 SMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRR 1257
Query: 1060 L 1060
L
Sbjct: 1258 L 1260
>TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Arabidopsis
thaliana}, partial (23%)
Length = 1254
Score = 503 bits (1295), Expect = e-142
Identities = 250/289 (86%), Positives = 270/289 (92%)
Frame = +3
Query: 908 KELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSIT 967
KELVMLPL+RPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT+AGANFINISMSSIT
Sbjct: 3 KELVMLPLKRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSIT 182
Query: 968 SKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDG 1027
SKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDG
Sbjct: 183 SKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDG 362
Query: 1028 LRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDV 1087
LRTK+ ER++VLAATNRP+DLDEAVIRRLPRRLMV+LPDAPNR KIL+VILAKEDL++DV
Sbjct: 363 LRTKEKERILVLAATNRPFDLDEAVIRRLPRRLMVDLPDAPNRGKILRVILAKEDLAADV 542
Query: 1088 DLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIR 1147
DL AIANMTDGYSGSDLKNLCVTAAH PI+EILEKEKK+ + A+AE +P P L S DIR
Sbjct: 543 DLEAIANMTDGYSGSDLKNLCVTAAHCPIREILEKEKKDKSLALAENKPEPELCSSADIR 722
Query: 1148 SLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
L MEDF++AH+QVCASVSSES NM EL QWN+LYGEGGSR K+LSYF
Sbjct: 723 PLKMEDFRYAHEQVCASVSSESTNMNELQQWNDLYGEGGSRKMKSLSYF 869
>TC91833 similar to GP|20259341|gb|AAM13995.1 unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 1313
Score = 390 bits (1002), Expect = e-108
Identities = 203/215 (94%), Positives = 204/215 (94%), Gaps = 1/215 (0%)
Frame = +1
Query: 404 AACFIHLKHKEHAKYTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQL 463
AACFIHLKHKEHAKYTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQL
Sbjct: 1 AACFIHLKHKEHAKYTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQL 180
Query: 464 LLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTDPPASETDTPSSSNVPTPLGL 523
LLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTDPPASETDTPSSSNVPTPLGL
Sbjct: 181 LLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTDPPASETDTPSSSNVPTPLGL 360
Query: 524 ESQAKLETDSVPSTSGTAKNCLFKLGDRVKYSSSSACLYQTSSSRYKGPSNGSRGKVVLI 583
ESQAKLETDSVPSTSGTAKNCLFKLGDRVKYSSSSACLYQTSSSR GPSNGSRGKVVLI
Sbjct: 361 ESQAKLETDSVPSTSGTAKNCLFKLGDRVKYSSSSACLYQTSSSR--GPSNGSRGKVVLI 534
Query: 584 FDDNPLSKIGVRFDKPIP-DGVDLGSACEAGQGFF 617
FDDNPLSKIGVRFDKPIP G+ LGSA G F
Sbjct: 535 FDDNPLSKIGVRFDKPIP*WGLILGSAW*GGSRIF 639
Score = 249 bits (636), Expect(3) = 7e-98
Identities = 129/161 (80%), Positives = 134/161 (83%), Gaps = 1/161 (0%)
Frame = +3
Query: 603 GVDLGSAC-EAGQGFFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEA 661
GVDLG + F + + + L+ LFEVVTSESRDSPFILFMKEA
Sbjct: 594 GVDLGKCLVRRVKDFSATSLIFDWKTAALTNSTNRLLIXLFEVVTSESRDSPFILFMKEA 773
Query: 662 EKSIVGNGDPYSFKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDL 721
EKSIVGNGDPYSFKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDL
Sbjct: 774 EKSIVGNGDPYSFKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDL 953
Query: 722 AFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEAL 762
AFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEAL
Sbjct: 954 AFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEAL 1076
Score = 124 bits (311), Expect(3) = 7e-98
Identities = 61/62 (98%), Positives = 61/62 (98%)
Frame = +1
Query: 763 LASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKILGWA 822
LASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENS KILGWA
Sbjct: 1078 LASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSXKILGWA 1257
Query: 823 LS 824
LS
Sbjct: 1258 LS 1263
Score = 25.4 bits (54), Expect(3) = 7e-98
Identities = 11/17 (64%), Positives = 13/17 (75%)
Frame = +2
Query: 824 SHHLMQNPEADADAKLV 840
+HH MQN EA A+A LV
Sbjct: 1262 AHHXMQNXEAXANAXLV 1312
>TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22 {Arabidopsis
thaliana}, partial (39%)
Length = 1514
Score = 379 bits (973), Expect = e-105
Identities = 214/511 (41%), Positives = 313/511 (60%), Gaps = 29/511 (5%)
Frame = +1
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHT 692
+K +++L++V+ S S IL++K EK +G+ Y F+ L KL +V+++GS
Sbjct: 85 EKLFLHSLYKVLVSISXAGSVILYIKNVEKVFLGSPRMYRLFQKTLNKLSGSVLILGS-- 258
Query: 693 HSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKV 752
R +D K N+ LT LFP +
Sbjct: 259 ------------------------------------RPYDLKYNCTKVNEKLTMLFPYNI 330
Query: 753 TIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTN 812
I PQDE L WK QL + ++ +K H+ VL+ + + D L+++ D+T+ +
Sbjct: 331 EITPPQDETHLKIWKSQLKKAMKKTHLKDYTTHIAEVLAANDLYCDDLDTVDHNDMTILS 510
Query: 813 ENSEKILGWALSHHLM--QNPEADADAKLVLSSESIQYGIGIFQAIQNESKSLKK----- 865
+E+++ A+ HHL +NP+ + L++S++S+++ + +FQ ++ K KK
Sbjct: 511 NQTEEVVASAIFHHLKDAKNPKY-RNGILIISAKSLRHVLSLFQEGESSEKDNKKTKKES 687
Query: 866 ---------------------SLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVK 904
S K +N FE+ + ++IP N+I VTF DIGAL++VK
Sbjct: 688 KRDDSRKEKPKESKKDGDIKASAKSDSPDNAFEECIRQELIPANEIKVTFSDIGALDDVK 867
Query: 905 DTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMS 964
++L+E VMLPL+RP+LF + KPCKG+LLFGPPGTGKTMLAKA+A +AGA+FIN+S S
Sbjct: 868 ESLQEAVMLPLRRPDLFKGDGVLKPCKGVLLFGPPGTGKTMLAKAIANEAGASFINVSPS 1047
Query: 965 SITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVN 1024
+ITS W G+ EK V+A+FSLA+K+AP++IF+DEVDSMLG+R + EH +MR++KNEFM
Sbjct: 1048 TITSMWQGQSEKNVRALFSLAAKVAPTIIFIDEVDSMLGQRSSTREHSSMRRVKNEFMSR 1227
Query: 1025 WDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS 1084
WDGL +K E++ VLAATN P+DLDEA+IRR RR+MV LP A NR ILK ILAKE S
Sbjct: 1228 WDGLLSKPDEKITVLAATNMPFDLDEAIIRRFQRRIMVGLPSADNRETILKTILAKEK-S 1404
Query: 1085 SDVDLGAIANMTDGYSGSDLKNLCVTAAHRP 1115
+++ ++ MT+GYSGSDLKNLC+TAA+RP
Sbjct: 1405 ENMNFEELSTMTEGYSGSDLKNLCMTAAYRP 1497
>TC88876 similar to PIR|G96537|G96537 hypothetical protein F2J10.1 [imported]
- Arabidopsis thaliana, partial (72%)
Length = 1572
Score = 348 bits (894), Expect(2) = e-103
Identities = 194/456 (42%), Positives = 294/456 (63%), Gaps = 12/456 (2%)
Frame = +2
Query: 638 INTLFEVVTSESRDSPFILFMKEAEKSI---VGNGDPYSFKSKLEKLPDN-----VVVIG 689
+ L EV+ S+ P I++ ++ + + V + F K+E++ D V++ G
Sbjct: 152 VEALCEVLNSKR---PLIVYFPDSSQWLHKSVPKSNRNEFFHKVEEMFDRLYGPVVLICG 322
Query: 690 SH-THSDSRKEKSHAGGLLFTKFGSNQTALLDLA-FPDSFGRLHDRGKEVPKPNKTLTKL 747
+ HS S++++ ++ FG L L D F +G + + + + KL
Sbjct: 323 QNKVHSGSKEKEKFT--MILPNFGRVAKLPLSLKHLTDGF-----KGGKTSEEDD-INKL 478
Query: 748 FPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKD 807
F N +++H P++E L +K+QL+ D + + + NL+ LR VL + L +
Sbjct: 479 FSNVLSVHPPKEENLQTVFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCTDLLHVNTDG 658
Query: 808 LTLTNENSEKILGWALSHHLMQN--PEADADAKLVLSSESIQYGIGIFQAIQNESKSLKK 865
+ +T + +EK++GWA +H+L P + +L + ES++ I + ++ S+ +
Sbjct: 659 IVITKQKAEKLVGWAKNHYLSSCLLPSIKGE-RLCIPRESLEIAISRMKGMETMSRKSSQ 835
Query: 866 SLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQ 925
+LK++ ++EFE + V+ P +IGV FDDIGALE+VK L+ELV+LP++RPELF G
Sbjct: 836 NLKNLA-KDEFESNFVSAVVAPGEIGVKFDDIGALEDVKKALQELVILPMRRPELFSHGN 1012
Query: 926 LTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLA 985
L +PCKGILLFGPPGTGKT+LAKA+AT+AGANFI+++ S++TSKWFG+ EK KA+FS A
Sbjct: 1013 LLRPCKGILLFGPPGTGKTLLAKALATEAGANFISVTGSTLTSKWFGDAEKLTKALFSFA 1192
Query: 986 SKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRP 1045
SK+AP +IFVDEVDS+LG R EHEA R+M+NEFM WDGLR+K+ +R+++L ATNRP
Sbjct: 1193 SKLAPVIIFVDEVDSLLGARGGAHEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRP 1372
Query: 1046 YDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKE 1081
+DLD+AVIRRLPRR+ V+LPD NR KIL++ LAKE
Sbjct: 1373 FDLDDAVIRRLPRRIYVDLPDVENRMKILRIFLAKE 1480
Score = 45.1 bits (105), Expect(2) = e-103
Identities = 21/38 (55%), Positives = 27/38 (70%)
Frame = +1
Query: 1075 KVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAA 1112
K I K +L+ D +A++T+GYSGSDLKNLCV AA
Sbjct: 1459 KNISCKRNLNPDFQYDKLASLTEGYSGSDLKNLCVAAA 1572
>TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22 {Arabidopsis
thaliana}, partial (74%)
Length = 2270
Score = 206 bits (524), Expect(3) = 9e-88
Identities = 116/250 (46%), Positives = 160/250 (63%), Gaps = 60/250 (24%)
Frame = +1
Query: 816 EKILGWALSHHLMQNPEAD-ADAKLVLSSESIQYGIGIF---------------QAIQNE 859
+++L A+SHHLM+N + + + KL++S S+ + +GIF QA+++E
Sbjct: 1297 KRLLVSAISHHLMKNKDLEYRNGKLIISCNSLSHALGIFRKGKFSGRDTSKLEDQALKSE 1476
Query: 860 SKSLKKSL--------------------------------------------KDVVTENE 875
S+ +++ +V +NE
Sbjct: 1477 IPSVVETVAKPAVKAESAAPADGAAPVKKAEAEISTSVAKAGAEKPVAASKASEVPPDNE 1656
Query: 876 FEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILL 935
FEKR+ +VIP N+I VTF DIGALE K++L+ELVMLPL+RP+LF G L KPC+GILL
Sbjct: 1657 FEKRIRPEVIPANEIDVTFSDIGALEETKESLQELVMLPLRRPDLFTGG-LLKPCRGILL 1833
Query: 936 FGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFV 995
FGPPGTGKTMLAKA+A +AGA+FIN+SMS+ITSKWFGE EK V+A+F+LASK++P++IFV
Sbjct: 1834 FGPPGTGKTMLAKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFV 2013
Query: 996 DEVDSMLGRR 1005
DEVDSMLG+R
Sbjct: 2014 DEVDSMLGQR 2043
Score = 100 bits (248), Expect(3) = 9e-88
Identities = 46/72 (63%), Positives = 58/72 (79%)
Frame = +3
Query: 1009 GEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAP 1068
G +AMRK+KNEFM +WDGL TK ER++VLAATN+P+DLDEA+IRR RR+MV LP
Sbjct: 2055 GSMKAMRKIKNEFMSHWDGLTTKQGERILVLAATNKPFDLDEAIIRRFERRIMVGLPSVE 2234
Query: 1069 NRAKILKVILAK 1080
NR KIL+ +L+K
Sbjct: 2235 NREKILRTLLSK 2270
Score = 97.1 bits (240), Expect = 4e-20
Identities = 50/103 (48%), Positives = 68/103 (65%)
Frame = +2
Query: 358 STSGTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAK 417
S S +V + + I+DG+E V+FD FPYYLSE T+ +L +A ++HLKH E +K
Sbjct: 221 SFSSNAVTADKIEHEMLRQIVDGRESNVTFDKFPYYLSEQTRVLLTSAAYVHLKHAEVSK 400
Query: 418 YTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFD 460
YT +L + ILLSGPA E+Y ++L KALA YF AKLL+ D
Sbjct: 401 YTRNLAPASRTILLSGPA--ELYQQVLAKALAHYFEAKLLLLD 523
Score = 58.9 bits (141), Expect(3) = 9e-88
Identities = 36/149 (24%), Positives = 72/149 (48%), Gaps = 1/149 (0%)
Frame = +2
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHT 692
+K LI +L++V+ S S+ P +L++++ ++ + + Y+ F+ L++L ++++GS
Sbjct: 860 EKLLIQSLYKVLLSVSKTYPIVLYLRDVDRLLSRSQKIYNMFQKMLKRLSGPILILGS-- 1033
Query: 693 HSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKV 752
R+ D G E + ++ LT LFP +
Sbjct: 1034------------------------------------RVLDSGNEFEELDEKLTVLFPYDI 1105
Query: 753 TIHMPQDEALLASWKQQLDRDVETLKIKG 781
I P+DE+ L SWK QL+ D++ +++ G
Sbjct: 1106KIRPPEDESHLVSWKSQLEEDMKMIQVSG 1192
Score = 33.1 bits (74), Expect = 0.66
Identities = 40/168 (23%), Positives = 69/168 (40%), Gaps = 8/168 (4%)
Frame = +2
Query: 849 GIGIFQAIQNESK--SLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGAL--ENVK 904
GIG+ + QN SK + VT ++ E +L ++ + VTFD E +
Sbjct: 167 GIGLANSGQNVSKWGPNYSFSSNAVTADKIEHEMLRQIVDGRESNVTFDKFPYYLSEQTR 346
Query: 905 DTLKELVMLPLQRPELFCKGQLTKPC-KGILLFGPPGTGKTMLAKAVATDAGANFINISM 963
L + L+ E+ + P + ILL GP + +LAKA+A A + + +
Sbjct: 347 VLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVLAKALAHYFEAKLLLLDL 526
Query: 964 SSITSK---WFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP 1008
+ + K +G G K S + + + S+ +RE P
Sbjct: 527 TDFSLKIQGKYGTGNKEFSFKRSTSESTLEKLSDIFGSFSIFSQREEP 670
>TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regulatory
particle chain RPT6-like protein {Arabidopsis thaliana},
partial (77%)
Length = 1564
Score = 286 bits (732), Expect = 3e-77
Identities = 144/271 (53%), Positives = 193/271 (71%)
Frame = +1
Query: 854 QAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVML 913
+A++ + + K+ + ++ N +E + DVI P+ I V FD IG LE +K TL ELV+L
Sbjct: 283 KALEQKKEIAKRLGRPLIQTNSYEDVIACDVINPDHIDVEFDSIGGLETIKQTLFELVIL 462
Query: 914 PLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGE 973
PLQRP+LF G+L P KG+LL+GPPGTGKTMLAKA+A ++GA FIN+ +S++ SKWFG+
Sbjct: 463 PLQRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFINVRISNLMSKWFGD 642
Query: 974 GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDT 1033
+K V AVFSLA K+ PS+IF+DEVDS LG+R + +HEA+ MK EFM WDG T +
Sbjct: 643 AQKLVAAVFSLAHKLQPSIIFIDEVDSFLGQRRS-SDHEAVLNMKTEFMALWDGFATDQS 819
Query: 1034 ERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIA 1093
RV+VLAATNRP +LDEA++RRLP+ + PD RA ILKVIL E + ++D IA
Sbjct: 820 ARVMVLAATNRPSELDEAILRRLPQAFEIGYPDRKERADILKVILKGEKVEDNIDFSYIA 999
Query: 1094 NMTDGYSGSDLKNLCVTAAHRPIKEILEKEK 1124
+ GY+GSDL +LC AA+ PI+EIL EK
Sbjct: 1000 GLCKGYTGSDLFDLCKKAAYFPIREILHNEK 1092
>AW584618 similar to GP|22831135|d putative MSP1(mitochondrial sorting of
proteins) protein homolog, partial (7%)
Length = 676
Score = 199 bits (507), Expect = 4e-51
Identities = 99/99 (100%), Positives = 99/99 (100%)
Frame = +1
Query: 1098 GYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHA 1157
GYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHA
Sbjct: 7 GYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHA 186
Query: 1158 HQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
HQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF
Sbjct: 187 HQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 303
>TC89617 similar to GP|22831135|dbj|BAC15996. putative MSP1(mitochondrial
sorting of proteins) protein homolog, partial (11%)
Length = 1062
Score = 190 bits (482), Expect = 3e-48
Identities = 93/125 (74%), Positives = 109/125 (86%)
Frame = +2
Query: 1072 KILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAV 1131
KI++VILAKE+L+ DVDL A+ANMTDGYSGSDLKNLCVTAAH PI+EILEKEKKE +A+
Sbjct: 56 KIMRVILAKEELAPDVDLEALANMTDGYSGSDLKNLCVTAAHCPIREILEKEKKERTSAL 235
Query: 1132 AEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKK 1191
AE +P P L S DIR L +EDFK+AH+QVCASVSS+S NMTEL+QWN+LYGEGGSR K
Sbjct: 236 AENKPLPRLCSSADIRPLKIEDFKYAHEQVCASVSSDSTNMTELLQWNDLYGEGGSRKKT 415
Query: 1192 ALSYF 1196
+LSYF
Sbjct: 416 SLSYF 430
Score = 38.1 bits (87), Expect = 0.020
Identities = 20/40 (50%), Positives = 26/40 (65%)
Frame = +1
Query: 1054 RRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIA 1093
RRLPRRLMVNLPDA NR K + +K + + G+I+
Sbjct: 1 RRLPRRLMVNLPDAANREKNYESHFSKRRIGA*C*FGSIS 120
>TC89903 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22 {Arabidopsis
thaliana}, partial (20%)
Length = 875
Score = 187 bits (474), Expect = 3e-47
Identities = 98/189 (51%), Positives = 130/189 (67%), Gaps = 10/189 (5%)
Frame = +1
Query: 1018 KNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVI 1077
KNEFM +WDGL + E+++VL ATNRP+DLDEA+IRR RR+MV LP A NR ILK +
Sbjct: 1 KNEFMTHWDGLLSGPNEQILVLGATNRPFDLDEAIIRRFERRIMVGLPSAENREMILKTL 180
Query: 1078 LAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKK---ELAAAVAEG 1134
++KE ++D +A MT+G+SGSDLKNLC+TAA+RP++E++++E+K E AE
Sbjct: 181 MSKEK-HENLDFKELATMTEGFSGSDLKNLCITAAYRPVRELIQQERKKEMEKKKKEAES 357
Query: 1135 RPAPALRGSDD-------IRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGS 1187
+ S+D +R L+MED K A QV AS +SE M EL QWNELYGEGGS
Sbjct: 358 VNSEDASNSNDNDEPEIILRPLSMEDMKQAKNQVAASFASEGSVMAELKQWNELYGEGGS 537
Query: 1188 RVKKALSYF 1196
R K+ L+YF
Sbjct: 538 RKKQQLTYF 564
>TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, partial (74%)
Length = 1193
Score = 182 bits (462), Expect = 7e-46
Identities = 97/219 (44%), Positives = 145/219 (65%), Gaps = 1/219 (0%)
Frame = +3
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V ++D+ LE+ K L+E V+LP++ P+ F + +P + LL+GPPGTGK+ LAKAVA
Sbjct: 51 VKWNDVAGLESAKQALQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 224
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
T+A + F +IS S + SKW GE EK V +F +A + APS+IFVDE+DS+ G+R E
Sbjct: 225 TEADSTFFSISSSDLVSKWMGESEKLVSNLFQMARESAPSIIFVDEIDSLCGQRGEGNES 404
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRA 1071
EA R++K E +V G+ D ++V+VLAATN PY LD+A+ RR +R+ + LPD R
Sbjct: 405 EASRRIKTELLVQMQGVGNND-QKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQ 581
Query: 1072 KILKVILAKEDLS-SDVDLGAIANMTDGYSGSDLKNLCV 1109
+ KV L + ++ D +A+ T+G+SGSD+ ++CV
Sbjct: 582 HMFKVHLGDTPHNLTEKDYEYLASRTEGFSGSDI-SVCV 695
>TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, complete
Length = 1793
Score = 180 bits (457), Expect = 3e-45
Identities = 95/228 (41%), Positives = 144/228 (62%), Gaps = 1/228 (0%)
Frame = +2
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V ++D+ LE+ K +L+E V+LP++ P+ F + +P + LL+GPPGTGK+ LAKAVA
Sbjct: 569 VKWNDVAGLESAKQSLQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 742
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
T+A + F ++S S + SKW GE EK V +F +A + APS+IFVDE+DS+ G R E
Sbjct: 743 TEADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESAPSIIFVDEIDSLCGTRGEGNES 922
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRA 1071
EA R++K E +V G+ D ++V+VLAATN PY LD+A+ RR +R+ + LPD R
Sbjct: 923 EASRRIKTELLVQMQGVGHND-QKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQ 1099
Query: 1072 KILKVILAKEDLS-SDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE 1118
+ KV L + ++ D +A T+G+SGSD+ P+++
Sbjct: 1100 HMFKVHLGDTPHNLTESDFEHLARKTEGFSGSDIAVCVKDVLFEPVRK 1243
>TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle protein 48
homolog (Valosin containing protein homolog) (VCP).
[Soybean], partial (95%)
Length = 2785
Score = 177 bits (448), Expect = 3e-44
Identities = 105/282 (37%), Positives = 160/282 (56%), Gaps = 4/282 (1%)
Frame = +2
Query: 893 TFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT 952
++DDIG LENVK L+E V P++ PE F K ++ P KG+L +GPPG GKT+LAKA+A
Sbjct: 1529 SWDDIGGLENVKRELQETVQYPVEHPEKFEKFGMS-PSKGVLFYGPPGCGKTLLAKAIAN 1705
Query: 953 DAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP--GE 1010
+ ANFI+I + + WFGE E V+ +F A AP V+F DE+DS+ +R +
Sbjct: 1706 ECQANFISIKGPELLTMWFGESEANVREIFDKARGSAPCVLFFDELDSIATQRGSSVGDA 1885
Query: 1011 HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAP 1068
A ++ N+ + DG+ K T V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 1886 GGAADRVLNQLLTEMDGMSAKKT--VFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDED 2059
Query: 1069 NRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELA 1128
+R +I K L K +S DVD+ A+A T G+SG+D+ +C A I+E +EK+ E
Sbjct: 2060 SRHQIFKACLRKSPISKDVDIRALAKYTQGFSGADITEICQRACKYAIRENIEKD-IEKE 2236
Query: 1129 AAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESV 1170
+E A D++ + F+ + + SVS +
Sbjct: 2237 RKRSENPEAMEEDIEDEVAEIKAAHFEESMKYARRSVSDADI 2362
Score = 167 bits (422), Expect = 3e-41
Identities = 96/232 (41%), Positives = 142/232 (60%), Gaps = 2/232 (0%)
Frame = +2
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V +DD+G + ++ELV LPL+ P+LF K KP KGILL+GPPG+GKT++A+AVA
Sbjct: 707 VGYDDVGGVRKQMAQIRELVELPLRHPQLF-KSIGVKPPKGILLYGPPGSGKTLIARAVA 883
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+ GA F I+ I SK GE E ++ F A K APS+IF+DE+DS+ +RE
Sbjct: 884 NETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HG 1060
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPN 1069
E R++ ++ + DGL+++ VIV+ ATNRP +D A+ R R R + + +PD
Sbjct: 1061 EVERRIVSQLLTLMDGLKSR--AHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEVG 1234
Query: 1070 RAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILE 1121
R ++L++ L+ DVDL I+ T GY G+DL LC AA + I+E ++
Sbjct: 1235 RLEVLRIHTKNMKLAEDVDLEKISKETHGYVGADLAALCTEAALQCIREKMD 1390
>AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33019 [imported]
- Arabidopsis thaliana, partial (34%)
Length = 556
Score = 166 bits (421), Expect = 4e-41
Identities = 85/174 (48%), Positives = 116/174 (65%), Gaps = 6/174 (3%)
Frame = +3
Query: 891 GVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAV 950
GV +DD+ L K L+E V+LPL PE F + +P KG+L+FGPPGTGKT+LAKAV
Sbjct: 39 GVRWDDVAGLTEAKRLLEEAVVLPLWMPEYF--QGIRRPWKGVLMFGPPGTGKTLLAKAV 212
Query: 951 ATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGE 1010
AT+ G F N+S +++ SKW GE E+ V+ +F LA APS IF+DE+DS+ R GE
Sbjct: 213 ATECGTTFFNVSSATLASKWRGESERMVRCLFDLARAYAPSTIFIDEIDSLCNARGASGE 392
Query: 1011 HEAMRKMKNEFMVNWDGLRTKDTER------VIVLAATNRPYDLDEAVIRRLPR 1058
HE+ R++K+E +V DG+ T V+VLAATN P+D+DEA+ RRL +
Sbjct: 393 HESSRRVKSELLVQVDGVSNSATNEDGSRKLVMVLAATNFPWDIDEALRRRLEK 554
>TC87818 similar to GP|20197082|gb|AAC26698.2 putative katanin {Arabidopsis
thaliana}, partial (62%)
Length = 984
Score = 154 bits (389), Expect = 2e-37
Identities = 80/190 (42%), Positives = 122/190 (64%), Gaps = 1/190 (0%)
Frame = +2
Query: 938 PPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDE 997
P G + MLAKAVAT+ F NIS SS+ SKW G+ EK VK +F LA AP+ IF+DE
Sbjct: 5 PQGLERPMLAKAVATECNTTFFNISASSMFSKWRGDSEKLVKVLFELARHHAPATIFLDE 184
Query: 998 VDSMLGRR-ENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRL 1056
+D+++ +R E EHEA R++K E ++ DGL D E V VLAATN P++LD A++RRL
Sbjct: 185 IDAIISQRGEGRSEHEASRRLKTELLIQMDGLARTD-ELVFVLAATNLPWELDAAMLRRL 361
Query: 1057 PRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPI 1116
+R++V LP+ R + + +L + + + + T+GYSGSD++ LC A +P+
Sbjct: 362 EKRILVPLPEPEARRAMFEELLPLQPDEEPMPYDLLVDRTEGYSGSDIRLLCKETAMQPL 541
Query: 1117 KEILEKEKKE 1126
+ ++ + ++E
Sbjct: 542 RRLMTQLEQE 571
>TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AAA-ATPase
subunit RPT4a-like protein {Arabidopsis thaliana},
complete
Length = 1640
Score = 150 bits (380), Expect = 2e-36
Identities = 84/231 (36%), Positives = 140/231 (60%), Gaps = 5/231 (2%)
Frame = +2
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
+++ +G L + L+E + LPL PELF + + KP KG+LL+GPPGTGKT+LA+A+A
Sbjct: 476 ISYSAVGGLSDQIRELRESIELPLMNPELFLRVGI-KPPKGVLLYGPPGTGKTLLARAIA 652
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPG-- 1009
++ ANF+ + S+I K+ GE + ++ +F A P +IF+DE+D++ GRR + G
Sbjct: 653 SNIDANFLKVVSSAIIDKYIGESARLIREMFGYARDHQPCIIFMDEIDAIGGRRFSEGTS 832
Query: 1010 -EHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPD 1066
+ E R + E + DG +V ++ ATNRP LD A++R RL R++ + LP+
Sbjct: 833 ADREIQRTLM-ELLNQLDGF--DQLGKVKMIMATNRPDVLDPALLRPGRLDRKIEIPLPN 1003
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIK 1117
+R +ILK+ A ++D A+ + +G++G+DL+N+C A I+
Sbjct: 1004 EQSRMEILKIHAAGIAKHGEIDYEAVVKLAEGFNGADLRNVCTEAGMSAIR 1156
>TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidopsis thaliana},
partial (98%)
Length = 1633
Score = 147 bits (371), Expect = 2e-35
Identities = 89/232 (38%), Positives = 137/232 (58%), Gaps = 6/232 (2%)
Frame = +1
Query: 893 TFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT 952
T+D IG L+ +KE++ LP++ PELF + +P KG+LL+GPPGTGKT+LA+AVA
Sbjct: 616 TYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQP-KGVLLYGPPGTGKTLLARAVAH 792
Query: 953 DAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRR----ENP 1008
FI +S S + K+ GEG + V+ +F +A + APS+IF+DE+DS+ R
Sbjct: 793 HTDCTFIRVSGSELVQKYIGEGSRMVRELFVMAREHAPSIIFMDEIDSIGSARMESGSGN 972
Query: 1009 GEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPD 1066
G+ E R M E + DG + ++ VL ATNR LD+A++R R+ R++ P+
Sbjct: 973 GDSEVQRTML-ELLNQLDGFEA--SNKIKVLMATNRIDILDQALLRPGRIDRKIEFPNPN 1143
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE 1118
+R ILK+ + +L +DL IA +G SG++LK +C A ++E
Sbjct: 1144 EESRRDILKIHSRRMNLMRGIDLKKIAEKMNGASGAELKAVCTEAGMFALRE 1299
>TC76904 homologue to GP|4325041|gb|AAD17230.1| FtsH-like protein Pftf
precursor {Nicotiana tabacum}, partial (91%)
Length = 2667
Score = 145 bits (367), Expect = 7e-35
Identities = 92/227 (40%), Positives = 131/227 (57%), Gaps = 5/227 (2%)
Frame = +1
Query: 891 GVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAV 950
GVTFDD+ ++ K E+V L++PE F P KG+LL GPPGTGKT+LAKA+
Sbjct: 817 GVTFDDVAGVDEAKQDFMEVVEF-LKKPERFTSVGARIP-KGVLLVGPPGTGKTLLAKAI 990
Query: 951 ATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP-- 1008
A +AG F +IS S + G G V+ +F A + AP ++FVDE+D+ +GR+
Sbjct: 991 AGEAGVPFFSISGSEFVEMFVGIGASRVRDLFKKAKENAPCIVFVDEIDA-VGRQRGTGI 1167
Query: 1009 -GEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
G ++ + N+ + DG + VIV+AATNR LD A++R R R++ V++P
Sbjct: 1168 GGGNDEREQTLNQLLTEMDGF--EGNTGVIVVAATNRADILDSALLRPGRFDRQVSVDVP 1341
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAA 1112
D R +ILKV + +DV L IA T G+SG+DL NL AA
Sbjct: 1342 DVRGRTEILKVHANNKKFDNDVSLEVIAMRTPGFSGADLANLLNEAA 1482
>TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15.70 -
Arabidopsis thaliana, complete
Length = 1629
Score = 145 bits (365), Expect = 1e-34
Identities = 85/231 (36%), Positives = 135/231 (57%), Gaps = 5/231 (2%)
Frame = +2
Query: 893 TFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT 952
++ DIG L+ +KE V LPL PEL+ + KP KG++L+G PGTGKT+LAKAVA
Sbjct: 647 SYADIGGLDAQIQEIKEAVELPLTHPELY-EDIGIKPPKGVILYGEPGTGKTLLAKAVAN 823
Query: 953 DAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRR---ENPG 1009
A F+ + S + K+ G+G K V+ +F +A ++PS++F+DE+D++ +R + G
Sbjct: 824 STSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGG 1003
Query: 1010 EHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
E E R M E + DG ++ +VI+ ATNR LD A++R R+ R++ LPD
Sbjct: 1004 EREIQRTML-ELLNQLDGFDSRGDVKVIL--ATNRIESLDPALLRPGRIDRKIEFPLPDI 1174
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE 1118
R +I + ++ L+ DV+L D +SG+D+K +C A ++E
Sbjct: 1175 KTRRRIFTIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRE 1327
>TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome regulatory
particle triple-A ATPase subunit2b {Oryza sativa
(japonica cultivar-group), partial (98%)
Length = 1696
Score = 144 bits (364), Expect = 2e-34
Identities = 84/231 (36%), Positives = 136/231 (58%), Gaps = 5/231 (2%)
Frame = +3
Query: 893 TFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT 952
++ DIG L+ +KE V LPL PEL+ + KP KG++L+G PGTGKT+LAKAVA
Sbjct: 645 SYADIGGLDAQIQEIKEAVELPLTHPELY-EDIGIKPPKGVILYGEPGTGKTLLAKAVAN 821
Query: 953 DAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRR---ENPG 1009
A F+ + S + K+ G+G K V+ +F +A ++P+++F+DE+D++ +R + G
Sbjct: 822 STSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPAIVFIDEIDAVGTKRYDAHSGG 1001
Query: 1010 EHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
E E R M E + DG ++ +VI+ ATNR LD A++R R+ R++ LPD
Sbjct: 1002 EREIQRTML-ELLNQLDGFDSRGDVKVIL--ATNRIESLDPALLRPGRIDRKIEFPLPDI 1172
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE 1118
R +I ++ ++ L+ DV+L D +SG+D+K +C A ++E
Sbjct: 1173 KTRRRIFQIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRE 1325
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,369,237
Number of Sequences: 36976
Number of extensions: 475262
Number of successful extensions: 2348
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 2189
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2281
length of query: 1196
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1089
effective length of database: 5,058,295
effective search space: 5508483255
effective search space used: 5508483255
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC141863.4


