

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
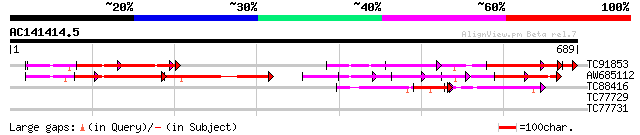
Query= AC141414.5 - phase: 0 /pseudo
(689 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91853 similar to GP|9955546|emb|CAC05431.1 putative protein {A... 184 4e-53
AW685112 similar to GP|9955546|emb putative protein {Arabidopsis... 184 8e-47
TC88416 similar to GP|21703136|gb|AAM74508.1 AT4g32250/F10M6_110... 63 3e-10
TC77729 weakly similar to GP|22037342|gb|AAM90012.1 disease resi... 39 0.007
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 39 0.007
>TC91853 similar to GP|9955546|emb|CAC05431.1 putative protein {Arabidopsis
thaliana}, partial (25%)
Length = 667
Score = 184 bits (468), Expect(2) = 4e-53
Identities = 90/92 (97%), Positives = 91/92 (98%)
Frame = +1
Query: 580 PSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRT 639
PSS+LERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKL IYTPAGSRT
Sbjct: 4 PSSYLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLRIYTPAGSRT 183
Query: 640 WMLDPSEVEVVEEKELCIGDWVRVRASVSTPT 671
WMLDPSEVEVVEEKELCIGDWVRVRASVSTPT
Sbjct: 184 WMLDPSEVEVVEEKELCIGDWVRVRASVSTPT 279
Score = 120 bits (302), Expect = 1e-27
Identities = 70/202 (34%), Positives = 111/202 (54%), Gaps = 7/202 (3%)
Frame = +1
Query: 457 VEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGD 516
+E+V VGQ VR + + +PRFGW G S G I I ADG++R+ S W D
Sbjct: 16 LERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLRIYTPAGSRTWMLD 195
Query: 517 PSDLQAEQIFE--VGEWVRLKENVN----NWKSIGPGSVGVVQGIGYEGGETDRSTFVGF 570
PS+++ + E +G+WVR++ +V+ +W + S+G V + D + +V F
Sbjct: 196 PSEVEVVEEKELCIGDWVRVRASVSTPTHHWGEVSHSSIGGVHRV------EDDNLWVSF 357
Query: 571 CGEQEKWVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLW 630
C + W+ +S +ERV VG KVR++ + PR+GW THAS G + +DA+GKL
Sbjct: 358 CFVERLWLCKASEMERVRPYKVGDKVRIRDGLVSPRWGWGMETHASRGHVVRVDANGKLR 537
Query: 631 I-YTPAGSRTWMLDPSEVEVVE 651
I + R W+ DP+++ + E
Sbjct: 538 IRFRWREGRPWIGDPADIALDE 603
Score = 105 bits (261), Expect = 8e-23
Identities = 55/128 (42%), Positives = 80/128 (61%), Gaps = 2/128 (1%)
Frame = +1
Query: 82 GQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEMERVE 141
GQ V++K +V +PRFGW G + SIGT+ +D DG LR+ P SR W DP+E+E VE
Sbjct: 43 GQKVRVKQNVKQPRFGWSGHTHASIGTIQAID-ADGKLRIYTPAGSRTWMLDPSEVEVVE 219
Query: 142 EFK--VGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCEPEE 199
E + +GDWVRVR +++T H G V +IG V+ + D +L V +V+ W C+ E
Sbjct: 220 EKELCIGDWVRVRASVSTPTHHWGEVSHSSIGGVHRVE-DDNLWVSFCFVERLWLCKASE 396
Query: 200 IEHVPPFR 207
+E V P++
Sbjct: 397 MERVRPYK 420
Score = 95.5 bits (236), Expect = 6e-20
Identities = 49/142 (34%), Positives = 80/142 (55%), Gaps = 3/142 (2%)
Frame = +1
Query: 386 WKVSPGDAERLPGXE--VGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELAC 443
W + P + E + E +GDWVR + S+ P++ W V S+ VH V+D L ++
Sbjct: 184 WMLDPSEVEVVEEKELCIGDWVRVRASVST-PTHHWGEVSHSSIGGVHRVEDDN-LWVSF 357
Query: 444 CFRKGKWITHYTDVEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVR 503
CF + W+ +++E+V +KVG VR R GL PR+GWG S+G + + A+G++R
Sbjct: 358 CFVERLWLCKASEMERVRPYKVGDKVRIRDGLVSPRWGWGMETHASRGHVVRVDANGKLR 537
Query: 504 VAFFGLSGL-WKGDPSDLQAEQ 524
+ F G W GDP+D+ ++
Sbjct: 538 IRFRWREGRPWIGDPADIALDE 603
Score = 72.4 bits (176), Expect = 5e-13
Identities = 56/193 (29%), Positives = 86/193 (44%), Gaps = 14/193 (7%)
Frame = +1
Query: 22 VKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVLTS---------- 71
V+ K+ V +P+ GW G S+G +Q++ G++R+ T
Sbjct: 52 VRVKQNVKQPRFGWSGHTHASIGTIQAIDA-----------DGKLRIYTPAGSRTWMLDP 198
Query: 72 ---EIVKLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASR 128
E+V+ L G V+++ V+ P W S SIG V V ED L V F R
Sbjct: 199 SEVEVVEEKELCIGDWVRVRASVSTPTHHWGEVSHSSIGGVHRV--EDDNLWVSFCFVER 372
Query: 129 GWKADPAEMERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSY 188
W +EMERV +KVGD VR+R L + + G G + G V + + L + +
Sbjct: 373 LWLCKASEMERVRPYKVGDKVRIRDGLVSPRWGWGMETHASRGHVVRVDANGKLRIRFRW 552
Query: 189 VQ-NPWHCEPEEI 200
+ PW +P +I
Sbjct: 553 REGRPWIGDPADI 591
Score = 62.4 bits (150), Expect = 6e-10
Identities = 42/121 (34%), Positives = 66/121 (53%), Gaps = 3/121 (2%)
Frame = +1
Query: 20 DWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVL--TSEIVKLI 77
DWV+ + +V+ P H W +S+G V V ++D+L VSFC E L SE+ ++
Sbjct: 241 DWVRVRASVSTPTHHWGEVSHSSIGGVHRV---EDDNLWVSFCFVERLWLCKASEMERVR 411
Query: 78 PLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFP-GASRGWKADPAE 136
P G V+++ + PR+GW ++ S G V+ VD +G LR+ F R W DPA+
Sbjct: 412 PYKVGDKVRIRDGLVSPRWGWGMETHASRGHVVRVD-ANGKLRIRFRWREGRPWIGDPAD 588
Query: 137 M 137
+
Sbjct: 589 I 591
Score = 42.7 bits (99), Expect(2) = 4e-53
Identities = 18/18 (100%), Positives = 18/18 (100%)
Frame = +3
Query: 672 PPLGGSESFKHRGGASGG 689
PPLGGSESFKHRGGASGG
Sbjct: 279 PPLGGSESFKHRGGASGG 332
Score = 35.4 bits (80), Expect = 0.074
Identities = 19/67 (28%), Positives = 30/67 (44%)
Frame = +1
Query: 137 MERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCE 196
+ERV++ VG VRV+ + + G +IG + I D L + W +
Sbjct: 16 LERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLRIYTPAGSRTWMLD 195
Query: 197 PEEIEHV 203
P E+E V
Sbjct: 196 PSEVEVV 216
Score = 31.6 bits (70), Expect = 1.1
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 1/76 (1%)
Frame = +1
Query: 383 QSLWKVSPGDAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELA 442
+ LW + ER+ +VGD VR + L + P + W S V V +G L +
Sbjct: 367 ERLWLCKASEMERVRPYKVGDKVRIRDGLVS-PRWGWGMETHASRGHVVRVDANGKLRIR 543
Query: 443 CCFRKGK-WITHYTDV 457
+R+G+ WI D+
Sbjct: 544 FRWREGRPWIGDPADI 591
>AW685112 similar to GP|9955546|emb putative protein {Arabidopsis thaliana},
partial (14%)
Length = 519
Score = 184 bits (468), Expect = 8e-47
Identities = 108/172 (62%), Positives = 108/172 (62%), Gaps = 40/172 (23%)
Frame = +1
Query: 189 VQNPWHCEPEEIEHVPPFR----------------------------------------N 208
VQNPWHCEPEEIEHVPPFR N
Sbjct: 1 VQNPWHCEPEEIEHVPPFRVRGM*IILIL*LNIFVLFLLHDLYLSYRLVIGYV*SDLLQN 180
Query: 209 LDMLGVVRHITVLEE*VRLRMMVS***KYQIDPYHGKLIHLIWRRWKILR*QFFWLMLII 268
LDMLGVVRHITVLEE*VRLRMM S***KYQIDPYHGKLIHLIWRRWKIL
Sbjct: 181 LDMLGVVRHITVLEE*VRLRMMXS***KYQIDPYHGKLIHLIWRRWKIL----------- 327
Query: 269 CFHFTEIRLEIGSE*KLQYLLQNMDGRILQGTVLV*FIAWKRMAIWVLPFAS 320
RLEIGS *KLQYLLQNMDGRILQGTVLV*FI RMAIWV PFAS
Sbjct: 328 -------RLEIGSX*KLQYLLQNMDGRILQGTVLV*FIXCXRMAIWVXPFAS 462
Score = 85.9 bits (211), Expect = 5e-17
Identities = 38/112 (33%), Positives = 71/112 (62%)
Frame = +3
Query: 79 LDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEME 138
L G V +K V EPR+ W G++ S+G + ++ DG+L + P W+ADP++ME
Sbjct: 135 LQIGDRVCVKRSVAEPRYAWGGETHHSVGRISEIE-NDGLLIIEIPNRPIPWQADPSDME 311
Query: 139 RVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQ 190
+VE+FKVGDWV V+ ++++ K+G ++ +IG+++ + D ++ V +++
Sbjct: 312 KVEDFKVGDWVXVKASVSSPKYGWEDITRNSIGVIHXL*XDGNMGVAFCFIK 467
Score = 77.4 bits (189), Expect = 2e-14
Identities = 38/81 (46%), Positives = 53/81 (64%)
Frame = +3
Query: 590 LIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTWMLDPSEVEV 649
L +G +V VK++V +PR+ W G TH S+G I I+ DG L I P W DPS++E
Sbjct: 135 LQIGDRVCVKRSVAEPRYAWGGETHHSVGRISEIENDGLLIIEIPNRPIPWQADPSDMEK 314
Query: 650 VEEKELCIGDWVRVRASVSTP 670
VE+ + +GDWV V+ASVS+P
Sbjct: 315 VEDFK--VGDWVXVKASVSSP 371
Score = 66.2 bits (160), Expect = 4e-11
Identities = 33/100 (33%), Positives = 56/100 (56%), Gaps = 4/100 (4%)
Frame = +3
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQAE 523
++G V + +AEPR+ WGG S G I+ I DG + + W+ DPSD++
Sbjct: 138 QIGDRVCVKRSVAEPRYAWGGETHHSVGRISEIENDGLLIIEIPNRPIPWQADPSDMEKV 317
Query: 524 QIFEVGEWVRLKENVNN----WKSIGPGSVGVVQGIGYEG 559
+ F+VG+WV +K +V++ W+ I S+GV+ + +G
Sbjct: 318 EDFKVGDWVXVKASVSSPKYGWEDITRNSIGVIHXL*XDG 437
Score = 60.5 bits (145), Expect = 2e-09
Identities = 33/107 (30%), Positives = 53/107 (48%)
Frame = +3
Query: 400 EVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVEK 459
++GD V K S+ P Y W S+ + +++ G L + R W +D+EK
Sbjct: 138 QIGDRVCVKRSVAE-PRYAWGGETHHSVGRISEIENDGLLIIEIPNRPIPWQADPSDMEK 314
Query: 460 VPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAF 506
V FKVG +V + ++ P++GW S G+I + DG + VAF
Sbjct: 315 VEDFKVGDWVXVKASVSSPKYGWEDITRNSIGVIHXL*XDGNMGVAF 455
Score = 54.3 bits (129), Expect = 2e-07
Identities = 28/92 (30%), Positives = 45/92 (48%)
Frame = +3
Query: 356 GGQMNHQQLLEK*CELTWMVPSMVTGRQSLWKVSPGDAERLPGXEVGDWVRSKPSLGNRP 415
GG+ +H E ++ + R W+ P D E++ +VGDWV K S+ + P
Sbjct: 195 GGETHHSVGRISEIENDGLLIIEIPNRPIPWQADPSDMEKVEDFKVGDWVXVKASVSS-P 371
Query: 416 SYDWNSVGRESLAVVHSVQDSGYLELACCFRK 447
Y W + R S+ V+H + G + +A CF K
Sbjct: 372 KYGWEDITRNSIGVIHXL*XDGNMGVAFCFIK 467
Score = 48.5 bits (114), Expect = 8e-06
Identities = 28/93 (30%), Positives = 48/93 (51%), Gaps = 3/93 (3%)
Frame = +3
Query: 20 DWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEV---RVLTSEIVKL 76
D V KR+V EP++ W G +SVG + + +ND L++ + S++ K+
Sbjct: 147 DRVCVKRSVAEPRYAWGGETHHSVGRISEI---ENDGLLIIEIPNRPIPWQADPSDMEKV 317
Query: 77 IPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTV 109
G V +K V+ P++GW +R+SIG +
Sbjct: 318 EDFKVGDWVXVKASVSSPKYGWEDITRNSIGVI 416
Score = 47.0 bits (110), Expect = 2e-05
Identities = 31/111 (27%), Positives = 48/111 (42%), Gaps = 4/111 (3%)
Frame = +3
Query: 525 IFEVGEWVRLKENVNN----WKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKWVGP 580
+ ++G+ V +K +V W SVG + I E D + W
Sbjct: 132 VLQIGDRVCVKRSVAEPRYAWGGETHHSVGRISEI-----ENDGLLIIEIPNRPIPWQAD 296
Query: 581 SSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWI 631
S +E+V+ VG V VK +V P++GW T SIG I + DG + +
Sbjct: 297 PSDMEKVEDFKVGDWVXVKASVSSPKYGWEDITRNSIGVIHXL*XDGNMGV 449
Score = 38.1 bits (87), Expect = 0.011
Identities = 18/47 (38%), Positives = 27/47 (57%)
Frame = +3
Query: 16 FDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFC 62
F V DWV K +V+ PK+GW+ NS+G + + N + V+FC
Sbjct: 324 FKVGDWVXVKASVSSPKYGWEDITRNSIGVIHXL*XDGN--MGVAFC 458
>TC88416 similar to GP|21703136|gb|AAM74508.1 AT4g32250/F10M6_110 {Arabidopsis
thaliana}, partial (52%)
Length = 2146
Score = 63.2 bits (152), Expect = 3e-10
Identities = 46/131 (35%), Positives = 61/131 (46%), Gaps = 8/131 (6%)
Frame = +3
Query: 529 GEWVRLKENVNNWKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVD 588
G+WVR+K+ VG++ I G VGF G Q W G S LE +
Sbjct: 1257 GDWVRVKDEKEKHSP-----VGILHSINRNDGRAS----VGFIGLQTLWNGNPSELEMAE 1409
Query: 589 KLIVGQKVRVKQNVKQPRFGWSGHT--HASIGTIQAIDADGKLWIYTPA------GSRTW 640
VGQ VR K+N+ PRF W ++ G I I +G L + P S T+
Sbjct: 1410 SFCVGQFVRPKENLLSPRFEWRRKRGGASATGRISWILPNGCLVVKFPGMMSFGNESTTF 1589
Query: 641 MLDPSEVEVVE 651
+ DPSEVEVV+
Sbjct: 1590 LADPSEVEVVD 1622
Score = 46.6 bits (109), Expect = 3e-05
Identities = 23/49 (46%), Positives = 33/49 (66%), Gaps = 1/49 (2%)
Frame = +3
Query: 491 GIITNIHA-DGEVRVAFFGLSGLWKGDPSDLQAEQIFEVGEWVRLKENV 538
GI+ +I+ DG V F GL LW G+PS+L+ + F VG++VR KEN+
Sbjct: 1305 GILHSINRNDGRASVGFIGLQTLWNGNPSELEMAESFCVGQFVRPKENL 1451
Score = 42.7 bits (99), Expect = 5e-04
Identities = 41/154 (26%), Positives = 66/154 (42%), Gaps = 11/154 (7%)
Frame = +3
Query: 398 GXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSV-QDSGYLELACCFRKGKWITHYTD 456
G GDWVR K + ++HS+ ++ G + + W + ++
Sbjct: 1245 GLAAGDWVRVKDEKEKH----------SPVGILHSINRNDGRASVGFIGLQTLWNGNPSE 1394
Query: 457 VEKVPSFKVGQYVRFRPGLAEPRFGW----GGAQPESQGIITNIHADGEVRVAFFGL--- 509
+E SF VGQ+VR + L PRF W GGA + G I+ I +G + V F G+
Sbjct: 1395 LEMAESFCVGQFVRPKENLLSPRFEWRRKRGGA--SATGRISWILPNGCLVVKFPGMMSF 1568
Query: 510 ---SGLWKGDPSDLQAEQIFEVGEWVRLKENVNN 540
S + DPS+++ V ++V N
Sbjct: 1569 GNESTTFLADPSEVEVVDFNTCPGMVEKYQHVEN 1670
Score = 39.7 bits (91), Expect = 0.004
Identities = 18/59 (30%), Positives = 30/59 (50%)
Frame = +3
Query: 101 QSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEMERVEEFKVGDWVRVRPTLTTSK 159
+ +G + ++ DG VGF G W +P+E+E E F VG +VR + L + +
Sbjct: 1287 EKHSPVGILHSINRNDGRASVGFIGLQTLWNGNPSELEMAESFCVGQFVRPKENLLSPR 1463
Score = 32.0 bits (71), Expect = 0.81
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 8/68 (11%)
Frame = +3
Query: 82 GQHVQLKGDVNEPRFGWRGQ--SRDSIGTVLCVDPEDGILRVGFPG------ASRGWKAD 133
GQ V+ K ++ PRF WR + + G + + P +G L V FPG S + AD
Sbjct: 1422 GQFVRPKENLLSPRFEWRRKRGGASATGRISWILP-NGCLVVKFPGMMSFGNESTTFLAD 1598
Query: 134 PAEMERVE 141
P+E+E V+
Sbjct: 1599 PSEVEVVD 1622
>TC77729 weakly similar to GP|22037342|gb|AAM90012.1 disease resistance-like
protein GS3-5 {Glycine max}, partial (72%)
Length = 973
Score = 38.9 bits (89), Expect = 0.007
Identities = 23/82 (28%), Positives = 38/82 (46%)
Frame = +1
Query: 394 ERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITH 453
+ L G +V DW R N P+ D + + S + + S +L++ACCF+ KW
Sbjct: 82 DNLFGRKVEDWKRILDEYENIPNKDIQRILQVSYDALEPKEKSVFLDIACCFKGCKW--- 252
Query: 454 YTDVEKVPSFKVGQYVRFRPGL 475
T V+K+ G + G+
Sbjct: 253 -TKVKKILHAHYGHCIEHHVGV 315
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 38.9 bits (89), Expect = 0.007
Identities = 23/82 (28%), Positives = 38/82 (46%)
Frame = +1
Query: 394 ERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITH 453
+ L G +V DW R N P+ D + + S + + S +L++ACCF+ KW
Sbjct: 1192 DNLFGRKVEDWKRILDEYENIPNKDIQRILQVSYDALEPKEKSVFLDIACCFKGCKW--- 1362
Query: 454 YTDVEKVPSFKVGQYVRFRPGL 475
T V+K+ G + G+
Sbjct: 1363 -TKVKKILHAHYGHCIEHHVGV 1425
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.144 0.480
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,437,509
Number of Sequences: 36976
Number of extensions: 395716
Number of successful extensions: 2050
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 1974
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2035
length of query: 689
length of database: 9,014,727
effective HSP length: 103
effective length of query: 586
effective length of database: 5,206,199
effective search space: 3050832614
effective search space used: 3050832614
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141414.5


