

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
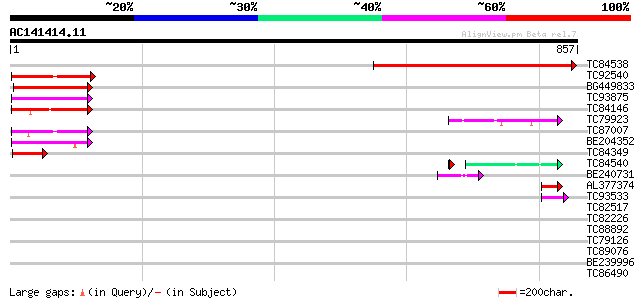
Query= AC141414.11 - phase: 0
(857 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to p... 549 e-156
TC92540 similar to GP|22136924|gb|AAM91806.1 putative RNA helica... 96 6e-20
BG449833 similar to GP|8096593|dbj Similar to Arabidopsis thalia... 93 5e-19
TC93875 similar to PIR|T49915|T49915 pre-mRNA splicing factor AT... 89 5e-18
TC84146 similar to GP|10440614|gb|AAG16852.1 putative ATP-depend... 88 2e-17
TC79923 similar to PIR|T04748|T04748 hypothetical protein T16H5.... 79 6e-15
TC87007 homologue to PIR|T48023|T48023 ATP-dependent RNA helicas... 75 8e-14
BE204352 similar to GP|1353239|gb putative RNA helicase A {Arabi... 66 5e-11
TC84349 similar to PIR|D86390|D86390 hypothetical protein AAF985... 55 1e-07
TC84540 similar to GP|21539577|gb|AAM53341.1 unknown protein {Ar... 50 9e-07
BE240731 weakly similar to PIR|E84647|E846 hypothetical protein ... 44 2e-04
AL377374 weakly similar to GP|8885575|dbj| emb|CAB75487.1~gene_i... 43 4e-04
TC93533 similar to GP|17065464|gb|AAL32886.1 Unknown protein {Ar... 42 8e-04
TC82517 homologue to GP|7688065|emb|CAB89694.1 constitutively ph... 35 0.094
TC82226 similar to PIR|T48615|T48615 hypothetical protein F18O22... 33 0.36
TC88892 similar to GP|4519673|dbj|BAA75685.1 WREBP-2 {Nicotiana ... 33 0.61
TC79126 similar to PIR|H96703|H96703 probable RING zinc finger p... 33 0.61
TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protei... 33 0.61
BE239996 similar to GP|21689819|gb| unknown protein {Arabidopsis... 32 1.0
TC86490 similar to PIR|T48615|T48615 hypothetical protein F18O22... 32 1.0
>TC84538 weakly similar to GP|22296418|dbj|BAC10186. similar to pre-mRNA
splicing factor ATP-dependent RNA helicase, partial
(11%)
Length = 973
Score = 549 bits (1414), Expect = e-156
Identities = 248/306 (81%), Positives = 275/306 (89%)
Frame = +2
Query: 551 KKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQVVKNFGPDLHGLKEKVPGADLRLNTRNR 610
+KLIQSLLSLH++KQ VI L G+DLPSD MKQVVKNFGPDLHGLKEKVPGADL LNTR +
Sbjct: 2 QKLIQSLLSLHEKKQLVISLRGKDLPSDLMKQVVKNFGPDLHGLKEKVPGADLELNTRQQ 181
Query: 611 TILCHGNSELKSRVEEITFEIARLSNPSSERFDTGPSCPICLCEVEDGYQLEGCGHLFCQ 670
I HGN ELK RVEEIT EIAR S+ ER DTGPSCPICLCEVEDGY+LEGCGHLFC+
Sbjct: 182 IIFLHGNKELKPRVEEITLEIARSSHHLVERLDTGPSCPICLCEVEDGYKLEGCGHLFCR 361
Query: 671 SCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSS 730
C+VEQCESAIKNQGSFPI CAHQGCG+ ILL DFRTLLSNDKL+ELFRASLGAFVASSS
Sbjct: 362 LCLVEQCESAIKNQGSFPICCAHQGCGDPILLTDFRTLLSNDKLDELFRASLGAFVASSS 541
Query: 731 GTYRFCPSPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQFKDDP 790
GTYRFCPSPDCPS+YRVAD DTAS PFVCGACYSETCT+CH+EYHPY+SCERYR+ KDDP
Sbjct: 542 GTYRFCPSPDCPSVYRVADSDTASEPFVCGACYSETCTKCHLEYHPYLSCERYRELKDDP 721
Query: 791 DSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIECKCGKHICWVCLEFFTTSGECYSHMD 850
DSSL++WCKGKEQVK+C ACG +IEK+DGCNH+ECKCGKH+CWVCLE FT+S ECY H+
Sbjct: 722 DSSLKEWCKGKEQVKSCFACGQIIEKIDGCNHVECKCGKHVCWVCLEIFTSSDECYDHLR 901
Query: 851 TIHLGV 856
TIH+ +
Sbjct: 902 TIHMTI 919
>TC92540 similar to GP|22136924|gb|AAM91806.1 putative RNA helicase
{Arabidopsis thaliana}, partial (24%)
Length = 836
Score = 95.9 bits (237), Expect = 6e-20
Identities = 53/129 (41%), Positives = 80/129 (61%), Gaps = 3/129 (2%)
Frame = +1
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESG---GCY 60
Q+ V++GETGSGK+TQ+ Q+L ++G + I CTQPRR+AA S+A RV E G G
Sbjct: 415 QVLVIVGETGSGKTTQIPQYLHEAGYTKHGMIACTQPRRVAAMSVAARVAHELGVKLGHE 594
Query: 61 EDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLL 120
SI+ F + + +MTD LL+ ++ + + S ++VDEAHER+++TD+L
Sbjct: 595 VGYSIR----FEDCTSDKTILKYMTDGMLLREFLGEPDLASYSVVMVDEAHERTLSTDIL 762
Query: 121 LALVPSSNR 129
ALV +R
Sbjct: 763 FALVKDISR 789
>BG449833 similar to GP|8096593|dbj Similar to Arabidopsis thaliana DNA
chromosome 4 BAC clone F28J12; RNA helicase - like
protein, partial (38%)
Length = 646
Score = 92.8 bits (229), Expect = 5e-19
Identities = 50/120 (41%), Positives = 76/120 (62%), Gaps = 1/120 (0%)
Frame = +3
Query: 6 TVLIGETGSGKSTQLVQFLADSG-VGANESIVCTQPRRIAAKSLAERVREESGGCYEDSS 64
T+++GETGSGK+TQ+ Q+L ++G I CTQPRR+A ++++ RV +E G D
Sbjct: 252 TIIVGETGSGKTTQIPQYLIEAGWASGGRLIACTQPRRLAVQAVSSRVAQEMGVKLGDQV 431
Query: 65 IKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
N+ ++ + F+TD LL+ M+D T S ++VDEAHERSI+TD+LL L+
Sbjct: 432 GYTIRFEDVTNQDETVLKFVTDGVLLREMMNDPLLTKYSVVMVDEAHERSISTDILLGLL 611
>TC93875 similar to PIR|T49915|T49915 pre-mRNA splicing factor ATP-dependent
RNA helicase-like protein - Arabidopsis thaliana,
partial (33%)
Length = 1239
Score = 89.4 bits (220), Expect = 5e-18
Identities = 49/121 (40%), Positives = 73/121 (59%)
Frame = +1
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ V++GETGSGK+TQL Q+L + G + CTQPRR+AA S+A+RV EE D
Sbjct: 280 QVVVVVGETGSGKTTQLTQYLYEDGYTIGGIVGCTQPRRVAAMSVAKRVSEEMDTELGD- 456
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F ++ I +MTD LL+ + D + I++DEAHERS++TD+L +
Sbjct: 457 KVGYAIRFEDVTGPNTVIKYMTDGVLLRETLKDSDLXKYRVIVMDEAHERSLSTDVLFGI 636
Query: 124 V 124
+
Sbjct: 637 L 639
>TC84146 similar to GP|10440614|gb|AAG16852.1 putative ATP-dependent RNA
helicase {Oryza sativa}, partial (24%)
Length = 797
Score = 87.8 bits (216), Expect = 2e-17
Identities = 46/125 (36%), Positives = 76/125 (60%), Gaps = 4/125 (3%)
Frame = +3
Query: 4 QITVLIGETGSGKSTQLVQFLADSGV----GANESIVCTQPRRIAAKSLAERVREESGGC 59
Q+ V+ GETG GK+TQL QF+ + + GA+ +I+CTQPRR++A S+A R+ E G
Sbjct: 102 QVLVVSGETGCGKTTQLPQFILEEEISCLRGADCNIICTQPRRVSAISVAARISAERGET 281
Query: 60 YEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
++ + + ++R++F T LL+ + D TG+S ++VDE HER +N D
Sbjct: 282 L-GKTVGYHIRLEAKRSAETRLLFCTTGVLLRQLVQDPELTGVSHLLVDEIHERGMNEDF 458
Query: 120 LLALV 124
L+ ++
Sbjct: 459 LIIIL 473
>TC79923 similar to PIR|T04748|T04748 hypothetical protein T16H5.30 -
Arabidopsis thaliana, partial (28%)
Length = 1001
Score = 79.3 bits (194), Expect = 6e-15
Identities = 50/185 (27%), Positives = 76/185 (41%), Gaps = 13/185 (7%)
Frame = +2
Query: 664 CGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLG 723
C H FC C+ + ++ PIRC GC I + ++ L E L +A
Sbjct: 326 CSHTFCSHCLRSYADGKLQCC-QVPIRCPQPGCRYCISTPECKSFLPFISFESLEKALSE 502
Query: 724 AFVASSSGTYRFCPSPDCP---------SIYRVADPDTASAPFVCGACYSETCTRCHIEY 774
A +A S Y CP P+C S + + ++ C C C C + +
Sbjct: 503 ANIAQSERFY--CPFPNCSVLLDPCECLSAMDGSSSQSDNSCIECPVCQRFICVGCGVPW 676
Query: 775 HPYVSCERYRQF----KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIECKCGKH 830
H +SCE ++ +D D +L + K K C C +IE GC H+ C+CG
Sbjct: 677 HSSMSCEEFQSLPEEERDASDITLHRLAQNKRW-KRCQQCRIMIELTQGCYHMTCRCGHE 853
Query: 831 ICWVC 835
C+ C
Sbjct: 854 FCYSC 868
>TC87007 homologue to PIR|T48023|T48023 ATP-dependent RNA helicase-like
protein - Arabidopsis thaliana, partial (96%)
Length = 2361
Score = 75.5 bits (184), Expect = 8e-14
Identities = 50/130 (38%), Positives = 73/130 (55%), Gaps = 9/130 (6%)
Frame = +3
Query: 4 QITVLIGETGSGKSTQLVQFLADS------GVGANESIVCTQPRRIAAKSLAERVREE-- 55
Q +L+GETGSGK+TQ+ QF+ ++ I CTQPRR+AA S++ RV EE
Sbjct: 393 QTLILVGETGSGKTTQIPQFVLEAVELEAPDKRKKMMIACTQPRRVAAMSVSRRVAEEMD 572
Query: 56 -SGGCYEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERS 114
S G SI+ F + + + ++TD LL+ M+D II+DEAHER+
Sbjct: 573 VSIGEEVGYSIR----FEDCSSARTVLKYLTDGMLLREAMTDPLLERYKVIILDEAHERT 740
Query: 115 INTDLLLALV 124
+ TD+L L+
Sbjct: 741 LATDVLFGLL 770
>BE204352 similar to GP|1353239|gb putative RNA helicase A {Arabidopsis
thaliana}, partial (13%)
Length = 572
Score = 66.2 bits (160), Expect = 5e-11
Identities = 47/134 (35%), Positives = 72/134 (53%), Gaps = 13/134 (9%)
Frame = +2
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANES--IVCTQPRRIAAKSLAERVREESGGCYE 61
Q+ ++ GETG GK+TQ+ Q++ D G E+ I+CTQPRRI+A S++ER+ E G
Sbjct: 14 QVVLISGETGCGKTTQVPQYIGDYMWGKGETCKILCTQPRRISAMSVSERISRERGEA-A 190
Query: 62 DSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSD------KN-----FTGISCIIVDEA 110
++ S S I+ T LL+ +S KN + I+ II+DE
Sbjct: 191 GENVGYKIRLDSKGGQQSSIVLCTTGVLLRVLVSKGSRRSMKNPAKDEISDITHIIMDEI 370
Query: 111 HERSINTDLLLALV 124
HER +D +LA++
Sbjct: 371 HERDRYSDFMLAIM 412
>TC84349 similar to PIR|D86390|D86390 hypothetical protein AAF98584.1
[imported] - Arabidopsis thaliana, partial (15%)
Length = 685
Score = 54.7 bits (130), Expect = 1e-07
Identities = 28/54 (51%), Positives = 40/54 (73%), Gaps = 1/54 (1%)
Frame = +2
Query: 5 ITVLIGETGSGKSTQLVQFLADSGVGANESIV-CTQPRRIAAKSLAERVREESG 57
I +++GETGSGK+TQ+ QFL +G + +V TQPRR+AA ++A+RV EE G
Sbjct: 389 ILIIVGETGSGKTTQIPQFLFHAGFCHDGKVVGITQPRRVAAITVAKRVAEECG 550
>TC84540 similar to GP|21539577|gb|AAM53341.1 unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 1027
Score = 49.7 bits (117), Expect(2) = 9e-07
Identities = 37/151 (24%), Positives = 60/151 (39%), Gaps = 4/151 (2%)
Frame = +3
Query: 689 IRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVA 748
+RC C + S + ++ R + +++ + T ++CP+P C
Sbjct: 588 LRCPDPACAAAVDQDMIDAFASAEDKKKYERYLVRSYIEVNKKT-KWCPAPGCEHAVNF- 761
Query: 749 DPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKN 806
D + C YS C C + H V C+ ++ K+ +S +W K
Sbjct: 762 DAGDENYDVSCLCSYS-FCWNCTEDAHRPVDCDTVSKWILKNSAESENTNWILA--YTKP 932
Query: 807 CPACGHVIEKVDGCNHIECK--CGKHICWVC 835
CP C IEK GC H+ C C CW+C
Sbjct: 933 CPKCKRSIEKNRGCMHMTCSAPCRFQFCWLC 1025
Score = 21.9 bits (45), Expect(2) = 9e-07
Identities = 6/9 (66%), Positives = 7/9 (77%)
Frame = +2
Query: 664 CGHLFCQSC 672
CGH +C SC
Sbjct: 509 CGHPYCSSC 535
>BE240731 weakly similar to PIR|E84647|E846 hypothetical protein At2g25360
[imported] - Arabidopsis thaliana, partial (18%)
Length = 603
Score = 44.3 bits (103), Expect = 2e-04
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Frame = +1
Query: 647 SCPICL--CEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVD 704
+C ICL +V + ++GC H +C SCM + E + + G PI C H+GC N +L+
Sbjct: 397 TCVICLEDSDVSQFFSVDGCQHRYCFSCMRQHVEVKLLH-GMVPI-CPHEGCKNELLVDS 570
Query: 705 FRTLLSNDKLE 715
L++ +E
Sbjct: 571 CPKFLTSKLVE 603
>AL377374 weakly similar to GP|8885575|dbj|
emb|CAB75487.1~gene_id:F15L12.13~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (7%)
Length = 386
Score = 43.1 bits (100), Expect = 4e-04
Identities = 14/31 (45%), Positives = 19/31 (61%)
Frame = +3
Query: 805 KNCPACGHVIEKVDGCNHIECKCGKHICWVC 835
K C C H+IE +GC H+ C+CG C+ C
Sbjct: 63 KQCVKCNHMIELAEGCYHMTCRCGFEFCYKC 155
>TC93533 similar to GP|17065464|gb|AAL32886.1 Unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 660
Score = 42.4 bits (98), Expect = 8e-04
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 2/42 (4%)
Frame = +1
Query: 805 KNCPACGHVIEKVDGCNHIEC--KCGKHICWVCLEFFTTSGE 844
K CP C IEK GC HI C C CW+CL ++ GE
Sbjct: 403 KPCPKCKRPIEKNQGCMHITCTPPCKFEFCWLCLGAWSDHGE 528
>TC82517 homologue to GP|7688065|emb|CAB89694.1 constitutively
photomorphogenic 1 protein {Pisum sativum}, partial
(22%)
Length = 671
Score = 35.4 bits (80), Expect = 0.094
Identities = 16/46 (34%), Positives = 26/46 (55%)
Frame = +3
Query: 648 CPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAH 693
CPIC+ ++D + L CGH FC C++ + ++N+ P C H
Sbjct: 159 CPICMQIIKDAF-LTSCGHSFCYMCII----THLRNKSDCPC-CGH 278
>TC82226 similar to PIR|T48615|T48615 hypothetical protein F18O22.210 -
Arabidopsis thaliana, partial (63%)
Length = 1300
Score = 33.5 bits (75), Expect = 0.36
Identities = 22/69 (31%), Positives = 28/69 (39%)
Frame = +3
Query: 604 RLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERFDTGPSCPICLCEVEDGYQLEG 663
R ++ N I NS +S + E + PSS CPICL +D G
Sbjct: 852 RASSSNAAISFRSNSFQQSTHTYASNENEMSTEPSSGGLYDNKVCPICLTNAKD--MAFG 1025
Query: 664 CGHLFCQSC 672
CGH C C
Sbjct: 1026CGHQTCCEC 1052
>TC88892 similar to GP|4519673|dbj|BAA75685.1 WREBP-2 {Nicotiana tabacum},
partial (90%)
Length = 1399
Score = 32.7 bits (73), Expect = 0.61
Identities = 26/84 (30%), Positives = 33/84 (38%)
Frame = +3
Query: 662 EGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRAS 721
EGCG F S +++ + + G C H+GCG L DF L S+ K
Sbjct: 435 EGCGKKFLDSSKLKR--HFLIHTGERDFVCPHEGCGKAFSL-DFN-LRSHMKTH------ 584
Query: 722 LGAFVASSSGTYRFCPSPDCPSIY 745
S Y CP PDC Y
Sbjct: 585 -------SQENYHICPYPDCGKRY 635
>TC79126 similar to PIR|H96703|H96703 probable RING zinc finger protein
T23K23.8 [imported] - Arabidopsis thaliana, partial (81%)
Length = 1397
Score = 32.7 bits (73), Expect = 0.61
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 2/30 (6%)
Frame = +3
Query: 648 CPICLCEVEDGYQLE--GCGHLFCQSCMVE 675
C ICLC EDG +L C H F SC+V+
Sbjct: 1104 CCICLCPYEDGVELHTLPCNHHFHSSCIVK 1193
>TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protein {Sorghum
bicolor}, partial (65%)
Length = 1326
Score = 32.7 bits (73), Expect = 0.61
Identities = 13/38 (34%), Positives = 21/38 (55%)
Frame = +2
Query: 635 SNPSSERFDTGPSCPICLCEVEDGYQLEGCGHLFCQSC 672
SN +++ + CP+CL +D GCGH+ C+ C
Sbjct: 1004 SNSTTDDQNQSACCPVCLTNAKD--LAFGCGHMTCRDC 1111
>BE239996 similar to GP|21689819|gb| unknown protein {Arabidopsis thaliana},
partial (16%)
Length = 420
Score = 32.0 bits (71), Expect = 1.0
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 9/57 (15%)
Frame = +2
Query: 625 EEITFEIARLSNPSSERFDT---------GPSCPICLCEVEDGYQLEGCGHLFCQSC 672
E I + A ++ P FD+ P CPICL +D GCGH C+ C
Sbjct: 128 EIIDHDNAFIATPRITNFDSIEPTAPASAEPVCPICLTNPKD--MAFGCGHTTCKEC 292
>TC86490 similar to PIR|T48615|T48615 hypothetical protein F18O22.210 -
Arabidopsis thaliana, partial (72%)
Length = 1739
Score = 32.0 bits (71), Expect = 1.0
Identities = 14/38 (36%), Positives = 18/38 (46%)
Frame = +3
Query: 635 SNPSSERFDTGPSCPICLCEVEDGYQLEGCGHLFCQSC 672
+ PS+ CPICL + +D GCGH C C
Sbjct: 1260 TEPSTSSLHDNKVCPICLTDAKD--MAFGCGHQTCCGC 1367
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,171,882
Number of Sequences: 36976
Number of extensions: 475492
Number of successful extensions: 2682
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 2626
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2668
length of query: 857
length of database: 9,014,727
effective HSP length: 104
effective length of query: 753
effective length of database: 5,169,223
effective search space: 3892424919
effective search space used: 3892424919
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC141414.11


