

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141111.8 - phase: 0 /pseudo
(779 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
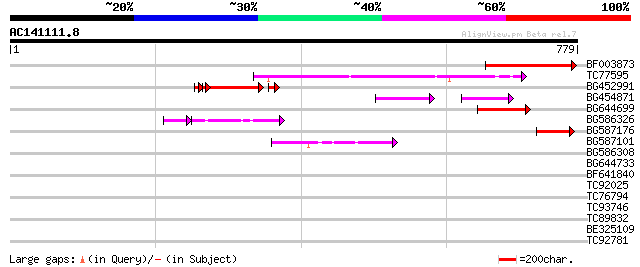
Score E
Sequences producing significant alignments: (bits) Value
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 228 7e-60
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 163 2e-40
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 109 2e-28
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 69 4e-15
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 63 5e-10
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 58 2e-09
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 51 1e-06
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 48 2e-05
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 37 0.022
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 36 0.049
BF641840 similar to GP|14334538|gb unknown protein {Arabidopsis ... 30 2.7
TC92025 similar to GP|10177284|dbj|BAB10637. gene_id:MJB21.19~pi... 30 3.5
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 30 3.5
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 30 3.5
TC89832 homologue to GP|21618319|gb|AAM67369.1 unknown {Arabidop... 30 3.5
BE325109 similar to PIR|T04011|T040 hypothetical protein T5L19.2... 30 4.6
TC92781 29 6.0
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 228 bits (581), Expect = 7e-60
Identities = 113/125 (90%), Positives = 117/125 (93%)
Frame = +2
Query: 654 GVGRALKSKKLTPKFIGPYHILERVGTVAYRVGLPPHLSNLHNVFHVSQLRKYVPDPSHV 713
GVGRALKSKKLT +FIGPY I ERVGTVAYRVGLPPHL NLH+VFHVSQLRKYVPDPSHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 714 IQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRVVWSGATGESLTWELESKMLES 773
IQSDDVQVRDNLTVETLP+RIDDRKVKTLRGKEIPLVRVVW A GESLTWELESKM+ES
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 774 YP*LF 778
YP LF
Sbjct: 362 YPELF 376
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 163 bits (413), Expect = 2e-40
Identities = 119/390 (30%), Positives = 189/390 (47%), Gaps = 14/390 (3%)
Frame = +2
Query: 335 KIDDQGVLRFRGRICIPDNE-------EIKKMILEESHRSSLSIHPGATKMYHDLKKIFW 387
++D L FRGRI +P ++ E++ +++ESH S+ + HPG + + F+
Sbjct: 83 QLDSLKRLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFF 262
Query: 388 GSGLKRDVAQFVYSCLVCQESKVEHQKPAGMMVPLDVLEWKWDSISMDFVTSLPNTP-RG 446
G + V +FV +C VC + Q G + PL V +SMDF+TSLP T RG
Sbjct: 263 WPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRG 442
Query: 447 SDAIWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFW 506
S +WVIVDRL+KS ++ + A+ ++ + HG+P SIVSDR + RFW
Sbjct: 443 SQYLWVIVDRLSKSVTLEEMD-TMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFW 619
Query: 507 KSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNN 566
+ G LS++YHPQTDG +ER Q ++ +LR V W LP ++ N
Sbjct: 620 REFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRN 799
Query: 567 SYHSSIGMASFEALYGRRCRTPLCWFESDERVVLGPE-----IVQQTTEKVQMIREKMKA 621
++SSIG F +G P+ E VV E +V++ + I+ ++ A
Sbjct: 800 RHNSSIGATPFFVEHGYHV-DPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVA 976
Query: 622 SQSRQKSYHDKRRKDLE-FKEGDHMFLRVTPMTGVGRALKSKKLTPKFIGPYHILERVGT 680
+Q R ++ +KRR + ++ GD ++L V+ + K L K Y + V
Sbjct: 977 AQQRSEASANKRRCPADRYQVGDKVWLNVSNYKSPRPSKKLDWLHHK----YEVTRFVTP 1144
Query: 681 VAYRVGLPPHLSNLHNVFHVSQLRKYVPDP 710
+ +P ++ FHV LR+ DP
Sbjct: 1145HVVELNVP---GTVYPKFHVDLLRRAASDP 1225
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 109 bits (272), Expect(4) = 2e-28
Identities = 54/78 (69%), Positives = 65/78 (83%)
Frame = +3
Query: 271 VREFELLEQFRDMSLVCEWTPQSVKLGMLKIDSEFLKSIKEAQRDDVKFVDLLVARDQTE 330
+ + LEQFRD+SLVCE +PQSVKLGMLKI++EFL SIKEAQ+ DVK VDL+ +QTE
Sbjct: 51 LESWSCLEQFRDLSLVCEVSPQSVKLGMLKINNEFLDSIKEAQKVDVKLVDLMFGNNQTE 230
Query: 331 DIDFKIDDQGVLRFRGRI 348
D DFK+DDQGVL+FR RI
Sbjct: 231 DGDFKVDDQGVLQFRDRI 284
Score = 26.9 bits (58), Expect(4) = 2e-28
Identities = 11/15 (73%), Positives = 15/15 (99%)
Frame = +2
Query: 356 IKKMILEESHRSSLS 370
+KKMILEESHRS+++
Sbjct: 284 MKKMILEESHRSNVN 328
Score = 25.4 bits (54), Expect(4) = 2e-28
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +1
Query: 254 VVADALSRKTLHM 266
VVAD LSRKTLH+
Sbjct: 1 VVADVLSRKTLHV 39
Score = 23.1 bits (48), Expect(4) = 2e-28
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = +2
Query: 265 HMSAMMVREFEL 276
+MSAMMVRE EL
Sbjct: 32 YMSAMMVRELEL 67
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 69.3 bits (168), Expect(2) = 4e-15
Identities = 36/81 (44%), Positives = 45/81 (55%)
Frame = +2
Query: 503 SRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEF 562
S FWK L + G+ L +SSAYHP +DGQSE + E LR + W P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 563 TYNNSYHSSIGMASFEALYGR 583
YN SY+ S M F+ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 30.4 bits (67), Expect(2) = 4e-15
Identities = 20/73 (27%), Positives = 34/73 (46%), Gaps = 2/73 (2%)
Frame = +1
Query: 622 SQSRQKSYHDKRRKDLEFKEGDHMFLRVTPMTGVGRALK--SKKLTPKFIGPYHILERVG 679
+Q K DK+R+ EF+ G+H+ +++ P AL+ K +P F +
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKYQKFGSPNFGSLLTVCSL*V 567
Query: 680 TVAYRVGLPPHLS 692
A+ PP+LS
Sbjct: 568 ESAFHCKSPPYLS 606
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 62.8 bits (151), Expect = 5e-10
Identities = 30/74 (40%), Positives = 49/74 (65%), Gaps = 1/74 (1%)
Frame = +2
Query: 643 DHMFLRVTPMT-GVGRALKSKKLTPKFIGPYHILERVGTVAYRVGLPPHLSNLHNVFHVS 701
+ + L+V P G R K KL+ ++IGP+ +++R+G VAY + LPP LS +H VFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 702 QLRKYVPDPSHVIQ 715
++Y D +++I+
Sbjct: 182 MFKRYHGDGNYIIR 223
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 57.8 bits (138), Expect(2) = 2e-09
Identities = 40/128 (31%), Positives = 70/128 (54%), Gaps = 1/128 (0%)
Frame = +2
Query: 251 KANVVADALSRKTLHMSAMMVREFELLEQFRDMSLVCEWTPQSVKLGMLKID-SEFLKSI 309
KAN+VADALSR+ + +SA RE + L+ + T + LG+ ++ ++ I
Sbjct: 359 KANLVADALSRRRVDVSAE--READDLDGMVRALRLNVLTKATESLGLEAVNQADLFTRI 532
Query: 310 KEAQRDDVKFVDLLVARDQTEDIDFKIDDQGVLRFRGRICIPDNEEIKKMILEESHRSSL 369
+ AQ D + + D+TE ++ G + GRI +P++ +K+ I+ E+H+S
Sbjct: 533 RLAQGQDEN-LQKVAQNDRTE---YQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRF 700
Query: 370 SIHPGATK 377
S+HPGA +
Sbjct: 701 SVHPGAPR 724
Score = 23.1 bits (48), Expect(2) = 2e-09
Identities = 17/38 (44%), Positives = 18/38 (46%)
Frame = +3
Query: 212 QDS*ELMRRIILRMISSWRP*SLC*RYGDIICMVRGLR 249
Q S*E MR MI W * *R+G CMV R
Sbjct: 126 QGS*ENMRETTPPMI*KWLR*YSP*RFGAHTCMVPRFR 239
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 51.2 bits (121), Expect = 1e-06
Identities = 23/51 (45%), Positives = 33/51 (64%)
Frame = -1
Query: 725 LTVETLPLRIDDRKVKTLRGKEIPLVRVVWSGATGESLTWELESKMLESYP 775
L +ET P+RI DR K +R K I +V++VW + E +TWE E++M YP
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYP 565
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 47.8 bits (112), Expect = 2e-05
Identities = 42/179 (23%), Positives = 84/179 (46%), Gaps = 6/179 (3%)
Frame = +2
Query: 360 ILEESHRSSLSIHPGATKMYHDLKKI-FWGSGLKRDVAQFVYSCLVCQES---KVEHQKP 415
IL H S+ + H +K +++ FW + +D F+ C CQ ++ P
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMP 283
Query: 416 AGMMVPLDVLEWKWDSISMDFVTSLPNTPRGSDAIWVIVDRLTKSAHFL--PINISFPVA 473
++ ++V +D +DF+ P++ + I V VD ++K + P N + V
Sbjct: 284 QNFILEVEV----FDVWGIDFMGPFPSS-YNNKYILVAVDYVSKWVEAIASPTNDATVVV 448
Query: 474 QLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSE 532
++ + I GVP ++SD F ++ ++ L + G + ++++AYHPQ +S+
Sbjct: 449 KM---FKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQKAERSK 616
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 37.4 bits (85), Expect = 0.022
Identities = 21/71 (29%), Positives = 37/71 (51%)
Frame = -2
Query: 487 HGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICV 546
HG+P IV+D F S ++ E +L +S +PQ++GQ+E + + + D L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 547 LEQGGTWDSHL 557
+ G W L
Sbjct: 505 DLKKGCWADEL 473
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 36.2 bits (82), Expect = 0.049
Identities = 18/40 (45%), Positives = 25/40 (62%)
Frame = -3
Query: 448 DAIWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLH 487
++I V+VDRLTKS F+P S+ A I + EIV +H
Sbjct: 121 ESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>BF641840 similar to GP|14334538|gb unknown protein {Arabidopsis thaliana},
partial (26%)
Length = 654
Score = 30.4 bits (67), Expect = 2.7
Identities = 17/48 (35%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Frame = +1
Query: 473 AQLAEIYIKEIVKLHGVPS-SIVSDRDPRFTSRFWKSLQEALGSKLRL 519
+ L +I++ + L+G P IVSDR+P++ S W ++ G KLR+
Sbjct: 382 SSLKDIHVDVVCILNGKPVWIIVSDRNPKYIS--WNECHKSKGLKLRI 519
>TC92025 similar to GP|10177284|dbj|BAB10637.
gene_id:MJB21.19~pir||T26506~similar to unknown protein
{Arabidopsis thaliana}, partial (20%)
Length = 697
Score = 30.0 bits (66), Expect = 3.5
Identities = 20/80 (25%), Positives = 41/80 (51%)
Frame = +1
Query: 258 ALSRKTLHMSAMMVREFELLEQFRDMSLVCEWTPQSVKLGMLKIDSEFLKSIKEAQRDDV 317
+L +L S +V+++ + +D SL+ + P+ + D+ +L+S K+A V
Sbjct: 13 SLHSASLEESLRIVKDYLIAATAKDCSLMICFRPRKEEGSGSAYDTVYLESTKQAFDFKV 192
Query: 318 KFVDLLVARDQTEDIDFKID 337
F+DL + R + +K+D
Sbjct: 193 HFIDLDLKRLSKVEDYYKLD 252
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 30.0 bits (66), Expect = 3.5
Identities = 12/21 (57%), Positives = 17/21 (80%)
Frame = -2
Query: 692 SNLHNVFHVSQLRKYVPDPSH 712
SNL N+FHVS + YVP+P++
Sbjct: 1690 SNLINIFHVS*FKVYVPNPTN 1628
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 30.0 bits (66), Expect = 3.5
Identities = 20/84 (23%), Positives = 38/84 (44%), Gaps = 13/84 (15%)
Frame = +2
Query: 394 DVAQFVYSCLVCQESKVEHQKPAGMMVPLDVLEWK-------------WDSISMDFVTSL 440
D + ++ L E K E KP G + ++ + K W+ +++DF L
Sbjct: 350 DAIKIKFAPLPLNEGKEEF-KPLGFLADMEPFKDKTKLCLLRPVPKPPWEDVTIDFSLGL 526
Query: 441 PNTPRGSDAIWVIVDRLTKSAHFL 464
T + D+ V+ D+ ++ AHF+
Sbjct: 527 L*TQQLKDSKMVVGDKFSRMAHFI 598
>TC89832 homologue to GP|21618319|gb|AAM67369.1 unknown {Arabidopsis
thaliana}, partial (62%)
Length = 1347
Score = 30.0 bits (66), Expect = 3.5
Identities = 18/49 (36%), Positives = 23/49 (46%)
Frame = +1
Query: 485 KLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSER 533
+++G PS I R FW S+ LGS R Y P TDG + R
Sbjct: 727 QIYGPPSKINKARQRTNVQVFWGSVH-GLGSFCRRCYCYSPDTDGIARR 870
>BE325109 similar to PIR|T04011|T040 hypothetical protein T5L19.200 -
Arabidopsis thaliana, partial (9%)
Length = 430
Score = 29.6 bits (65), Expect = 4.6
Identities = 14/43 (32%), Positives = 25/43 (57%)
Frame = +1
Query: 507 KSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQ 549
KSLQ G++++L + P+ D ERT+Q D +I + ++
Sbjct: 79 KSLQTKTGARIQLIPQHLPEGDDSKERTVQVTGDKRQIEIAQE 207
>TC92781
Length = 808
Score = 29.3 bits (64), Expect = 6.0
Identities = 26/105 (24%), Positives = 47/105 (44%), Gaps = 5/105 (4%)
Frame = -2
Query: 527 TDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIG---MASFEALYGR 583
T+ +++++Q ED L + G W HLP +E + S +S G S G
Sbjct: 534 TNDGTQKSLQGAEDCLAVFF---HGYWKQHLPPLEISLGTSMGASRGNPVKTSLGVSRGT 364
Query: 584 RCRTPLCWFESDERVVLGPEIVQQTTEK--VQMIREKMKASQSRQ 626
R E + L P+++ ++ + V++ R K S+SR+
Sbjct: 363 SFRATCTPSEVSKVHPLNPQVMIKSCVRFCVELCRSKGLQSESRK 229
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.336 0.146 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,930,655
Number of Sequences: 36976
Number of extensions: 390413
Number of successful extensions: 2718
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 1486
Number of HSP's successfully gapped in prelim test: 138
Number of HSP's that attempted gapping in prelim test: 1206
Number of HSP's gapped (non-prelim): 1664
length of query: 779
length of database: 9,014,727
effective HSP length: 104
effective length of query: 675
effective length of database: 5,169,223
effective search space: 3489225525
effective search space used: 3489225525
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141111.8


