

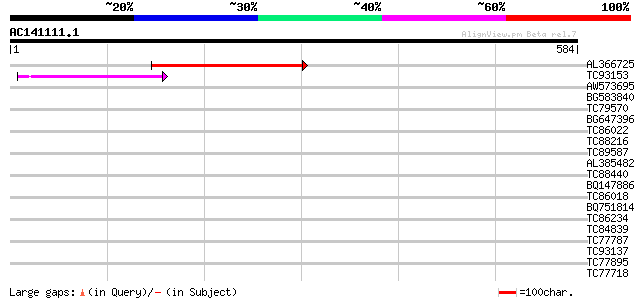
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141111.1 - phase: 0 /pseudo
(584 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AL366725 154 1e-37
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 75 5e-14
AW573695 weakly similar to GP|10177655|db gb|AAD43171.1~gene_id:... 35 0.061
BG583840 GP|20146556|gb ATP synthase alpha subunit {Clethra barb... 32 0.89
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 32 0.89
BG647396 weakly similar to PIR|T50791|T50 hypothetical protein T... 31 1.2
TC86022 homologue to GP|19423939|gb|AAL87312.1 putative RNA heli... 31 1.2
TC88216 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Pru... 30 2.6
TC89587 similar to PIR|C86306|C86306 prenyl transferase homolog ... 30 3.4
AL385482 similar to GP|5106924|gb|A putative cell wall protein {... 29 4.4
TC88440 weakly similar to GP|7293965|gb|AAF49324.1| Eip74EF gene... 29 4.4
BQ147886 weakly similar to GP|7242813|emb germin-like protein {P... 29 4.4
TC86018 similar to GP|19423939|gb|AAL87312.1 putative RNA helica... 29 4.4
BQ751814 similar to GP|18043248|gb| RIKEN cDNA 4833422F24 gene {... 29 4.4
TC86234 similar to SP|Q02758|ATPD_PEA ATP synthase delta chain ... 29 5.8
TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protei... 29 5.8
TC77787 29 5.8
TC93137 weakly similar to PIR|JQ2285|JQ2285 nodulin-26 - soybean... 29 5.8
TC77895 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 ... 29 5.8
TC77718 similar to GP|11072026|gb|AAG28905.1 F12A21.16 {Arabidop... 29 5.8
>AL366725
Length = 485
Score = 154 bits (388), Expect = 1e-37
Identities = 98/161 (60%), Positives = 113/161 (69%), Gaps = 1/161 (0%)
Frame = +3
Query: 147 SSTRTILLRMLSSRDVSSSRMV*GPTSRGRLGINS*EFF*IWLTAAGSMRRIPRLTTR** 206
SS RT+LLR+LSSR+ SSSRMV*GPTSRG+ NS EF I A S RRI RL TR *
Sbjct: 3 SSIRTMLLRLLSSRNASSSRMV*GPTSRGQSDTNSSEFSLI**IPAESTRRILRLMTRL* 182
Query: 207 MRGRARDSIVVLSRTVPPLTKGNKRWSMLGGLRRRM-LQRLCASIVVRKATRATPVLRKS 265
M GR R + V LS VP L + N+ WSM+G LRRRM L+RL S + RKATR VL++S
Sbjct: 183 MSGRPRANRVALSLIVPLLIRANREWSMIGVLRRRMLLRRLSVSTMARKATRVMFVLKRS 362
Query: 266 RSVSGVVRRVML*LIAIVQTLCALIAMERVTLVLNVLSLRG 306
R+VSGV RRV+L*LIA LCA IA +RV LV N SLRG
Sbjct: 363 RNVSGVTRRVIL*LIASEMILCASIATKRVILVPNASSLRG 485
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 75.5 bits (184), Expect = 5e-14
Identities = 61/154 (39%), Positives = 84/154 (53%)
Frame = +3
Query: 9 KLWDNNLT*MLVRMLKLGCWRRS*RRTLRLSKDVMTLMEPRRGLRRLRGFSGLCSARRIR 68
KL+++ L +LV +++ GC R S* + S+ M LM R G RRL+ +S C+ R R
Sbjct: 54 KLFNSYLRLILV-VMEPGC*RLS*GIIHQHSRVGMLLMVLRSG*RRLKEYSESCNVLRHR 230
Query: 69 KCGLVLISWPRKLMTGGLLFCLPLSRRELL*LGLFSGGSS*EDTFRKMFAGRRRSNSLS* 128
+C L ++ M GGL+ CL SR L * G SG S F++M R+R N S*
Sbjct: 231 RCSLGRTC*LKRPMIGGLVCCLFWSRMMLW*PGPCSGRSFWAGIFQRMSGVRKRLNFWS* 410
Query: 129 SKEICL*QSMLPSSWSCRSSTRTILLRMLSSRDV 162
++ CL QSML S W+ ST + R LSS V
Sbjct: 411 NRVTCLSQSMLQSLWNLPHSTLITVRRQLSSPSV 512
>AW573695 weakly similar to GP|10177655|db
gb|AAD43171.1~gene_id:MAC12.15~similar to unknown
protein {Arabidopsis thaliana}, partial (38%)
Length = 652
Score = 35.4 bits (80), Expect = 0.061
Identities = 26/83 (31%), Positives = 49/83 (58%), Gaps = 3/83 (3%)
Frame = +3
Query: 333 ILIILL*LLLLILVLRIAL--LLSIVFLLWVLSCLI*MERWLSKLQLRVR*LLL-SYV*N 389
+L+++L +LL ++++ +L LL ++ LLW + C +*+ WL L LR R ++L +
Sbjct: 204 MLLMVLKVLLNVILMSPSLERLLFLILLLWFMLCWL*LLCWLFMLLLRPRLVVLEEWDLT 383
Query: 390 VLCLCLVVILKWI*FVYL*VVWM 412
LC LV+ I ++ * +W+
Sbjct: 384 SLCPLLVLRRLRILIIFR*KLWL 452
>BG583840 GP|20146556|gb ATP synthase alpha subunit {Clethra barbinervis},
partial (17%)
Length = 585
Score = 31.6 bits (70), Expect = 0.89
Identities = 15/32 (46%), Positives = 24/32 (74%)
Frame = +2
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFLLWV 361
+L +L++LL LLLL+L+L + LLL I +W+
Sbjct: 11 LLLLLLLLLLLLLLLLLLLLLLLLYIYIYMWL 106
Score = 30.0 bits (66), Expect = 2.6
Identities = 14/29 (48%), Positives = 24/29 (82%)
Frame = +2
Query: 333 ILIILL*LLLLILVLRIALLLSIVFLLWV 361
+L++LL LLLL+L+L + LLL ++ LL++
Sbjct: 2 LLLLLLLLLLLLLLLLLLLLLLLLLLLYI 88
Score = 28.5 bits (62), Expect = 7.5
Identities = 13/31 (41%), Positives = 24/31 (76%)
Frame = +2
Query: 331 LAILIILL*LLLLILVLRIALLLSIVFLLWV 361
L +L++LL LLLL+L+L + LLL ++ +++
Sbjct: 2 LLLLLLLLLLLLLLLLLLLLLLLLLLLYIYI 94
Score = 28.5 bits (62), Expect = 7.5
Identities = 13/29 (44%), Positives = 24/29 (81%)
Frame = +2
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFL 358
+L +L++LL LLLL+L+L + LLL ++++
Sbjct: 2 LLLLLLLLLLLLLLLLLLLLLLLLLLLYI 88
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 31.6 bits (70), Expect = 0.89
Identities = 26/69 (37%), Positives = 43/69 (61%)
Frame = -3
Query: 334 LIILL*LLLLILVLRIALLLSIVFLLWVLSCLI*MERWLSKLQLRVR*LLLSYV*NVLCL 393
L++LL*LLLL+L++ + LLL ++ LL +L L+ M +S+ L ++ LL L
Sbjct: 667 LLLLL*LLLLLLLMLLLLLLLLLLLLLLL--LLHMIILISE*VLMLKKLL-------ELL 515
Query: 394 CLVVILKWI 402
CL + L+W+
Sbjct: 514 CLGLRLRWM 488
Score = 29.3 bits (64), Expect = 4.4
Identities = 14/30 (46%), Positives = 24/30 (79%)
Frame = -3
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFLL 359
+L +L++LL LLLL+L+L + LLL ++ L+
Sbjct: 646 LLLLLLMLLLLLLLLLLLLLLLLLHMIILI 557
Score = 28.5 bits (62), Expect = 7.5
Identities = 16/50 (32%), Positives = 33/50 (66%)
Frame = -3
Query: 331 LAILIILL*LLLLILVLRIALLLSIVFLLWVLSCLI*MERWLSKLQLRVR 380
L +L++L+ LLLL+L+L + LLL + ++ + ++ +++ L L L +R
Sbjct: 649 LLLLLLLMLLLLLLLLLLLLLLLLLHMIILISE*VLMLKKLLELLCLGLR 500
>BG647396 weakly similar to PIR|T50791|T50 hypothetical protein T30N20_90 -
Arabidopsis thaliana, partial (38%)
Length = 787
Score = 31.2 bits (69), Expect = 1.2
Identities = 25/72 (34%), Positives = 43/72 (59%), Gaps = 2/72 (2%)
Frame = +1
Query: 297 LVLNVLSLRGRRLLVGFSL*LGLRQKMRIV*SEVLAILIILL*LLLLILVLRIALLLSIV 356
L+L++L L GR LL LR ++R V +L++ + +LLL+++L + +L IV
Sbjct: 337 LILSLLELLGRCLL------RLLRWEVR*VWLLR*CVLLV*VLVLLLLMLLSMLVLQGIV 498
Query: 357 --FLLWVLSCLI 366
FL+W+L C +
Sbjct: 499 LRFLVWLLICRV 534
Score = 30.8 bits (68), Expect = 1.5
Identities = 45/143 (31%), Positives = 63/143 (43%), Gaps = 12/143 (8%)
Frame = +1
Query: 232 WSMLGGLRRRMLQRL--CASIVVRKATRATPVLRKSRSVSGVVRRVML*LIAIVQTLCAL 289
W ++GGL +L+ L C ++R R +LR * + +V L L
Sbjct: 319 WDIVGGLILSLLELLGRCLLRLLRWEVR*VWLLR--------------*CVLLV*VLVLL 456
Query: 290 IAMERVTLVLNVLSLRGR------RLLVGFSL*LGLRQKMRIV*SEVLAILIILL*LLLL 343
+ M LVL + LR R+ VGF L L VL ++I+L+ L +
Sbjct: 457 LLMLLSMLVLQGIVLRFLVWLLICRVCVGFVLVL------------VLFLVILLVVSLFI 600
Query: 344 ILVLRIALLLSIVFLL----WVL 362
ILVLR L+L FLL WVL
Sbjct: 601 ILVLRGRLVLWRFFLL*QLFWVL 669
>TC86022 homologue to GP|19423939|gb|AAL87312.1 putative RNA helicase
{Arabidopsis thaliana}, partial (89%)
Length = 1982
Score = 31.2 bits (69), Expect = 1.2
Identities = 23/51 (45%), Positives = 33/51 (64%)
Frame = -2
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFLLWVLSCLI*MERWLSKLQLRVR 380
+L +L+ILL +LLLIL++ + L+L V L +L LI M L K +RVR
Sbjct: 313 LLLLLLILLLILLLILIVLLMLILHHVPLHILLLLLILMMWSLLKPGVRVR 161
>TC88216 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Prunus
armeniaca}, partial (68%)
Length = 1508
Score = 30.0 bits (66), Expect = 2.6
Identities = 23/39 (58%), Positives = 26/39 (65%), Gaps = 5/39 (12%)
Frame = +3
Query: 333 ILIILL*LLLLILVLRIALLL----SIVFL-LWVLSCLI 366
IL+ILL L+LI +L LLL IVFL LWVLS LI
Sbjct: 564 ILVILLKELILIPILICMLLLLLIGGIVFLVLWVLSLLI 680
>TC89587 similar to PIR|C86306|C86306 prenyl transferase homolog [imported]
- Arabidopsis thaliana, partial (27%)
Length = 714
Score = 29.6 bits (65), Expect = 3.4
Identities = 13/23 (56%), Positives = 19/23 (82%)
Frame = +1
Query: 346 VLRIALLLSIVFLLWVLSCLI*M 368
V+R +L++IVFLLW+L L+*M
Sbjct: 265 VVRRGILINIVFLLWILLSLL*M 333
>AL385482 similar to GP|5106924|gb|A putative cell wall protein {Medicago
truncatula}, partial (42%)
Length = 402
Score = 29.3 bits (64), Expect = 4.4
Identities = 13/32 (40%), Positives = 26/32 (80%)
Frame = -2
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFLLWV 361
+L +L++++ LLL++L+L + LLL +V LL++
Sbjct: 143 LLVLLLVVVLLLLVVLLLVVVLLLLVVELLYL 48
>TC88440 weakly similar to GP|7293965|gb|AAF49324.1| Eip74EF gene product
{Drosophila melanogaster}, partial (5%)
Length = 711
Score = 29.3 bits (64), Expect = 4.4
Identities = 22/56 (39%), Positives = 37/56 (65%)
Frame = -3
Query: 329 EVLAILIILL*LLLLILVLRIALLLSIVFLLWVLSCLI*MERWLSKLQLRVR*LLL 384
E++ +L+++ *LLL++L L + LLL +V LL V+ L+ + L LQL + L+L
Sbjct: 364 EMMMVLLLVG*LLLVVL-LLVGLLLLVV*LLLVVLLLVGLLLVLHLLQLVLHLLVL 200
>BQ147886 weakly similar to GP|7242813|emb germin-like protein {Phaseolus
vulgaris}, partial (73%)
Length = 646
Score = 29.3 bits (64), Expect = 4.4
Identities = 12/27 (44%), Positives = 21/27 (77%)
Frame = +2
Query: 340 LLLLILVLRIALLLSIVFLLWVLSCLI 366
LLL++LV++I LL+ ++F +SCL+
Sbjct: 476 LLLVLLVVKIPLLIYLMFFCLAISCLL 556
>TC86018 similar to GP|19423939|gb|AAL87312.1 putative RNA helicase
{Arabidopsis thaliana}, partial (12%)
Length = 885
Score = 29.3 bits (64), Expect = 4.4
Identities = 13/30 (43%), Positives = 24/30 (79%)
Frame = -1
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFLL 359
+L +L+ILL +LLLIL++ + +LL ++ +L
Sbjct: 696 LLLLLLILLLILLLILLMMLIVLLMLILIL 607
>BQ751814 similar to GP|18043248|gb| RIKEN cDNA 4833422F24 gene {Mus
musculus}, partial (4%)
Length = 531
Score = 29.3 bits (64), Expect = 4.4
Identities = 22/56 (39%), Positives = 32/56 (56%), Gaps = 1/56 (1%)
Frame = +1
Query: 284 QTLCALIAMERVTLVLNVLSL-RGRRLLVGFSL*LGLRQKMRIV*SEVLAILIILL 338
Q L A +A L +L+L R +LVGFSL LGL R++ S +L +L++ L
Sbjct: 31 QKLAAKVAHRFFRLHTALLALLRSLLVLVGFSLGLGLFLLSRLLRSSLLQLLLVAL 198
>TC86234 similar to SP|Q02758|ATPD_PEA ATP synthase delta chain chloroplast
precursor (EC 3.6.3.14). [Garden pea] {Pisum sativum},
partial (98%)
Length = 1131
Score = 28.9 bits (63), Expect = 5.8
Identities = 27/64 (42%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Frame = +1
Query: 304 LRGR-RLLVGFSL*LGLRQKMRIV*SEVLAILIILL*LLLLILVLRIALLLSIVFLLWVL 362
LR R R L+G +* GL+ + V* VL +++L L LLI VLR L ++ L+WV+
Sbjct: 799 LRNRCRNLLGRRM*-GLKPILTRV*WRVLLSGMVILGLSLLI*VLRRNLRKLLLNLIWVI 975
Query: 363 SCLI 366
S L+
Sbjct: 976 SNLL 987
>TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protein~similar
to Arabidopsis thaliana chromosome 5 T6I14_10, partial
(20%)
Length = 819
Score = 28.9 bits (63), Expect = 5.8
Identities = 14/27 (51%), Positives = 22/27 (80%)
Frame = -1
Query: 330 VLAILIILL*LLLLILVLRIALLLSIV 356
+L +LI+LL LLLL+LVL + L+L ++
Sbjct: 246 LLVLLILLLLLLLLLLVLLVLLVLRVM 166
Score = 28.5 bits (62), Expect = 7.5
Identities = 16/37 (43%), Positives = 26/37 (70%)
Frame = -1
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFLLWVLSCLI 366
+L +L++ + LLL+LVL I LLL ++ LL +L L+
Sbjct: 288 LLLLLMVKMRRLLLLLVLLILLLLLLLLLLVLLVLLV 178
>TC77787
Length = 436
Score = 28.9 bits (63), Expect = 5.8
Identities = 19/45 (42%), Positives = 30/45 (66%), Gaps = 1/45 (2%)
Frame = +1
Query: 270 GVVRRVML*LIAIVQTLCALIAMERVTLVLNVLSLRG-RRLLVGF 313
G++ ++ +*+ IVQT+C L + +T NV SLRG R LL+G+
Sbjct: 61 GMI*KLNV*MQPIVQTMCILYFISALT--TNV*SLRGFRFLLIGY 189
>TC93137 weakly similar to PIR|JQ2285|JQ2285 nodulin-26 - soybean, partial
(47%)
Length = 633
Score = 28.9 bits (63), Expect = 5.8
Identities = 19/53 (35%), Positives = 31/53 (57%)
Frame = -2
Query: 340 LLLLILVLRIALLLSIVFLLWVLSCLI*MERWLSKLQLRVR*LLLSYV*NVLC 392
L L+L+L++A LL L L C++ +E + LQL ++*L Y+ +LC
Sbjct: 473 LACLLLILKLACLL----LSLKLVCMLILESYRQVLQLWLQ*LDQLYISTILC 327
>TC77895 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20
{Arabidopsis thaliana}, partial (41%)
Length = 2591
Score = 28.9 bits (63), Expect = 5.8
Identities = 34/121 (28%), Positives = 63/121 (51%), Gaps = 11/121 (9%)
Frame = +1
Query: 292 MERVTLVLNVLSLRGRRLLVGFSL*LGLRQKMRIV*SEVLAILIILL*LLLLILVLRIAL 351
+E V L+LN + L L++ + L L +++V + L + +ILL LLLL+L++ + +
Sbjct: 664 LELVILILNKM-LNSMVLVLWINKVLCLTLFLQMVTTITL*LALILLLLLLLLLLISLLV 840
Query: 352 ---------LLSIV--FLLWVLSCLI*MERWLSKLQLRVR*LLLSYV*NVLCLCLVVILK 400
L+S++ F ++ C I + ++L L +R L+S +* L L + L
Sbjct: 841 AEG*IKFKTLISVLHKFQFLMVVCFISINN--NQLGLLIRSYLISLL*TKTKLSLCLTLV 1014
Query: 401 W 401
W
Sbjct: 1015W 1017
>TC77718 similar to GP|11072026|gb|AAG28905.1 F12A21.16 {Arabidopsis
thaliana}, partial (14%)
Length = 1089
Score = 28.9 bits (63), Expect = 5.8
Identities = 20/46 (43%), Positives = 31/46 (66%), Gaps = 2/46 (4%)
Frame = +2
Query: 330 VLAILIILL*LLLLILVLRIALLLSIVFL--LWVLSCLI*MERWLS 373
++ I ILL L L++L+L++ LLL I+FL L++ LI + R LS
Sbjct: 344 LMFIFGILLVLFLVVLLLKLWLLLKIIFLANLFIRILLIEILRLLS 481
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.364 0.163 0.577
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,615,132
Number of Sequences: 36976
Number of extensions: 348133
Number of successful extensions: 5981
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 1997
Number of HSP's successfully gapped in prelim test: 278
Number of HSP's that attempted gapping in prelim test: 3561
Number of HSP's gapped (non-prelim): 2661
length of query: 584
length of database: 9,014,727
effective HSP length: 101
effective length of query: 483
effective length of database: 5,280,151
effective search space: 2550312933
effective search space used: 2550312933
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.5 bits)
S2: 61 (28.1 bits)
Medicago: description of AC141111.1


