

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
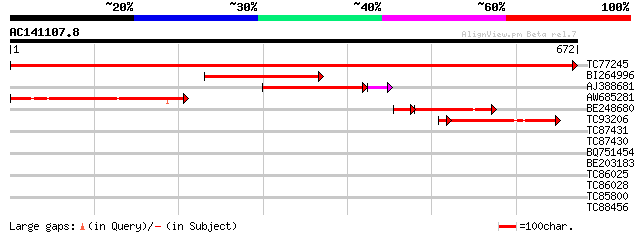
Query= AC141107.8 + phase: 0
(672 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77245 similar to GP|6630556|gb|AAF19575.1| putative alpha-L-ar... 1369 0.0
BI264996 weakly similar to GP|16417958|gb arabinosidase ARA-1 {L... 226 3e-59
AJ388681 similar to GP|13398412|gb arabinoxylan arabinofuranohyd... 207 4e-55
AW685281 weakly similar to GP|17380938|gb putative arabinosidase... 209 3e-54
BE248680 weakly similar to GP|13937191|gb AT3g10740/T7M13_18 {Ar... 132 6e-36
TC93206 similar to GP|13398412|gb|AAK21879.1 arabinoxylan arabin... 127 2e-31
TC87431 40 0.004
TC87430 40 0.004
BQ751454 weakly similar to GP|23326899|gb| alpha-L-arabinosidase... 35 0.093
BE203183 33 0.46
TC86025 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays... 30 3.9
TC86028 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays... 29 5.1
TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA b... 29 6.7
TC88456 similar to GP|15487292|dbj|BAB64531. vacuolar sorting re... 29 6.7
>TC77245 similar to GP|6630556|gb|AAF19575.1| putative
alpha-L-arabinofuranosidase {Arabidopsis thaliana},
partial (89%)
Length = 2351
Score = 1369 bits (3543), Expect = 0.0
Identities = 672/672 (100%), Positives = 672/672 (100%)
Frame = +2
Query: 1 MGFSKDSCCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFE 60
MGFSKDSCCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFE
Sbjct: 203 MGFSKDSCCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFE 382
Query: 61 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVAL 120
EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVAL
Sbjct: 383 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVAL 562
Query: 121 RLEVLCDGTCPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSL 180
RLEVLCDGTCPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSL
Sbjct: 563 RLEVLCDGTCPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSL 742
Query: 181 ASTVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHG 240
ASTVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHG
Sbjct: 743 ASTVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHG 922
Query: 241 FRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDD 300
FRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDD
Sbjct: 923 FRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDD 1102
Query: 301 GLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWG 360
GLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWG
Sbjct: 1103GLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWG 1282
Query: 361 SMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPL 420
SMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPL
Sbjct: 1283SMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPL 1462
Query: 421 DHPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAG 480
DHPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAG
Sbjct: 1463DHPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAG 1642
Query: 481 FLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGAT 540
FLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGAT
Sbjct: 1643FLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGAT 1822
Query: 541 LLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSS 600
LLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSS
Sbjct: 1823LLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSS 2002
Query: 601 GSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLKESSNLK 660
GSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLKESSNLK
Sbjct: 2003GSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLKESSNLK 2182
Query: 661 MLESDSSSWSSI 672
MLESDSSSWSSI
Sbjct: 2183MLESDSSSWSSI 2218
>BI264996 weakly similar to GP|16417958|gb arabinosidase ARA-1 {Lycopersicon
esculentum}, partial (20%)
Length = 422
Score = 226 bits (575), Expect = 3e-59
Identities = 95/140 (67%), Positives = 117/140 (82%)
Frame = +2
Query: 232 PLDTYKGHGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFG 291
PLDTYKGHGFR +L QM+ +LKP RFPGGC+VEG+ L+NAF+WK +GPWE RPGH+G
Sbjct: 2 PLDTYKGHGFRMNLFQMVAELKPRXFRFPGGCYVEGNVLKNAFQWKQTIGPWENRPGHYG 181
Query: 292 DVWKYWTDDGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFA 351
DVW YWTDDG G++E LQL+EDL ALPIWVFNNG+SH+++V+ SA+ P VQ ALDG+E A
Sbjct: 182 DVWDYWTDDGFGFFEGLQLAEDLNALPIWVFNNGISHSEQVNVSAISPSVQXALDGIEXA 361
Query: 352 RGDPTSKWGSMRAAMGHPEP 371
G PTS+WGS+ A MGHP+P
Sbjct: 362 IGSPTSRWGSIIAXMGHPKP 421
>AJ388681 similar to GP|13398412|gb arabinoxylan arabinofuranohydrolase
isoenzyme AXAH-I {Hordeum vulgare}, partial (18%)
Length = 574
Score = 207 bits (528), Expect(2) = 4e-55
Identities = 92/125 (73%), Positives = 110/125 (87%)
Frame = +1
Query: 300 DGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKW 359
DGLG++E LQL+ED+GALPIWVFNNG+SH+DEVDTS + PFV+EAL+G+EFARG TSKW
Sbjct: 1 DGLGFFEGLQLAEDIGALPIWVFNNGISHSDEVDTSVISPFVKEALEGIEFARGSSTSKW 180
Query: 360 GSMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRP 419
GS+RA+MGHP+PFNLKYVA+GNEDC KKNY GNY+ FY AI++ YPDIQIISNC P
Sbjct: 181 GSVRASMGHPKPFNLKYVAIGNEDCYKKNYYGNYMAFYKAIKKFYPDIQIISNCPAFKTP 360
Query: 420 LDHPA 424
L+HPA
Sbjct: 361 LNHPA 375
Score = 26.2 bits (56), Expect(2) = 4e-55
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Frame = +2
Query: 425 DMYDYHIY-TNANDMFSRSTTFNRVTRSGP 453
D+YDYH Y +A MF+ F++ GP
Sbjct: 377 DLYDYHTYPIDARXMFNAYHEFDKSPXHGP 466
>AW685281 weakly similar to GP|17380938|gb putative arabinosidase
{Arabidopsis thaliana}, partial (18%)
Length = 651
Score = 209 bits (532), Expect = 3e-54
Identities = 112/214 (52%), Positives = 146/214 (67%), Gaps = 3/214 (1%)
Frame = +2
Query: 1 MGFSKDSCCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFE 60
M FSK C L L+IV ++ QC ++LVV+A+Q GR +P TLFGIF+E
Sbjct: 23 MTFSKACCSFLWLYLIIVSFVAFQC---NANGSQISSLVVNAAQ--GRPMPNTLFGIFYE 187
Query: 61 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVAL 120
EINHAG+GG+WA+LV+N GFEA G PSNI PW+IIG + + ++T+ +SCFERNKVAL
Sbjct: 188 EINHAGSGGIWAQLVNNSGFEAAGTRTPSNIFPWTIIGTESSVKLQTELSSCFERNKVAL 367
Query: 121 RLEVLCDGTCPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSL 180
R++VLCD CP DGVGV NPGFWGMNI QGKKYKVVF+ RS G L+++V+ + L
Sbjct: 368 RMDVLCD-KCPPDGVGVSNPGFWGMNIVQGKKYKVVFFYRSLGSLDMRVAFRDAIXGRIL 544
Query: 181 ASTVI---TGSASDFSNWTKVETVLEAKATNPNS 211
AS+ I S V+T+LEA+A++ NS
Sbjct: 545 ASSHIIRHKASKKRVPKXQXVQTILEARASSXNS 646
>BE248680 weakly similar to GP|13937191|gb AT3g10740/T7M13_18 {Arabidopsis
thaliana}, partial (15%)
Length = 375
Score = 132 bits (332), Expect(2) = 6e-36
Identities = 64/98 (65%), Positives = 78/98 (79%)
Frame = +1
Query: 480 GFLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGA 539
GFLIGLEKNSD+V M +YAPLFVN NDR+W PDAIVF+S Q+YG PSYW+ F ES+GA
Sbjct: 76 GFLIGLEKNSDVVSMVNYAPLFVNTNDRKWTPDAIVFDSHQVYGIPSYWLIKLFKESSGA 255
Query: 540 TLLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVN 577
T LNS+LQT S +L ASAI+W++SVD + +RIK N
Sbjct: 256 TFLNSTLQTD-SPTLAASAISWKSSVDGTSILRIKVAN 366
Score = 37.4 bits (85), Expect(2) = 6e-36
Identities = 18/26 (69%), Positives = 21/26 (80%)
Frame = +3
Query: 456 FVSEYAVTGNDAGQGSLLAALAEAGF 481
FVSEYA+ DAG G+LLAA+AEA F
Sbjct: 3 FVSEYALIKEDAGNGTLLAAVAEARF 80
>TC93206 similar to GP|13398412|gb|AAK21879.1 arabinoxylan
arabinofuranohydrolase isoenzyme AXAH-I {Hordeum
vulgare}, partial (6%)
Length = 642
Score = 127 bits (319), Expect(2) = 2e-31
Identities = 69/133 (51%), Positives = 89/133 (66%)
Frame = +1
Query: 520 QLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFG 579
Q+YGTPSYW+ F ESNGAT LNS+LQTT +L ASAI ++ +K Y++IK N
Sbjct: 40 QVYGTPSYWVTYLFKESNGATFLNSTLQTTDPGTLAASAILVKDPQNKNTYLKIKIANMR 219
Query: 580 TSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMN 639
+ V+ KIS G +L+ GSTKTVLT N +DENSF++PKK+ P S LQ+ G +MN
Sbjct: 220 KTQVDFKISIQGFASKNLK--GSTKTVLTG-NELDENSFAEPKKIAPQTSPLQNPGNEMN 390
Query: 640 VIVPPHSFTSFDL 652
VIV P S T D+
Sbjct: 391 VIVQPTSLTILDM 429
Score = 27.3 bits (59), Expect(2) = 2e-31
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +2
Query: 509 WNPDAIVFNSFQLYG 523
WNPDAIVF+S + G
Sbjct: 8 WNPDAIVFSSIKFMG 52
>TC87431
Length = 662
Score = 39.7 bits (91), Expect = 0.004
Identities = 16/21 (76%), Positives = 18/21 (85%)
Frame = +2
Query: 261 GGCFVEGDYLRNAFRWKAAVG 281
GGCFVE +YLRN +WKAAVG
Sbjct: 437 GGCFVERNYLRNEIQWKAAVG 499
>TC87430
Length = 649
Score = 39.7 bits (91), Expect = 0.004
Identities = 16/21 (76%), Positives = 18/21 (85%)
Frame = +3
Query: 261 GGCFVEGDYLRNAFRWKAAVG 281
GGCFVE +YLRN +WKAAVG
Sbjct: 459 GGCFVERNYLRNEIQWKAAVG 521
>BQ751454 weakly similar to GP|23326899|gb| alpha-L-arabinosidase
{Bifidobacterium longum NCC2705}, partial (12%)
Length = 780
Score = 35.0 bits (79), Expect = 0.093
Identities = 17/48 (35%), Positives = 26/48 (53%)
Frame = +3
Query: 240 GFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERP 287
GFR D+++ + +L +R+PGG F + W VGP E+RP
Sbjct: 255 GFRKDVVEAVKELNCPVMRYPGGNFCA------TYHWIDGVGPREKRP 380
>BE203183
Length = 408
Score = 32.7 bits (73), Expect = 0.46
Identities = 12/17 (70%), Positives = 14/17 (81%)
Frame = +2
Query: 261 GGCFVEGDYLRNAFRWK 277
GGCFVE +YLRN +WK
Sbjct: 356 GGCFVERNYLRNEIQWK 406
>TC86025 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays}, partial
(90%)
Length = 1890
Score = 29.6 bits (65), Expect = 3.9
Identities = 13/35 (37%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Frame = +1
Query: 87 IPSNIDPW-SIIGNATYINVETDRTSCFERNKVAL 120
+P I+ W S GN TY+N+E + +S F+ V +
Sbjct: 1264 VPLTINCWPSSAGNETYVNIEYEASSMFDLRNVVI 1368
>TC86028 similar to GP|7677262|gb|AAF67098.1| delta-COP {Zea mays}, partial
(34%)
Length = 992
Score = 29.3 bits (64), Expect = 5.1
Identities = 13/35 (37%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Frame = +3
Query: 87 IPSNIDPW-SIIGNATYINVETDRTSCFERNKVAL 120
+P I+ W S GN TY+N+E + +S F+ V +
Sbjct: 204 VPLTINCWPSSSGNETYVNIEYEASSMFDLRNVVI 308
>TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA binding
protein {Oryza sativa (japonica cultivar-group)},
partial (37%)
Length = 2028
Score = 28.9 bits (63), Expect = 6.7
Identities = 21/80 (26%), Positives = 38/80 (47%)
Frame = -2
Query: 105 VETDRTSCFERNKVALRLEVLCDGTCPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGP 164
+E + S E + L++ C C +G G++ E+ V+F + + P
Sbjct: 428 LEVEGFSSVETISFLMMLDLTC-WLCSCEGDGLFLS-------EKKSTSPVIFRSPPSSP 273
Query: 165 LNLKVSLTGSNGVGSLASTV 184
L L+V LTG+ +G++ S V
Sbjct: 272 LVLRVRLTGNPRIGNILSLV 213
>TC88456 similar to GP|15487292|dbj|BAB64531. vacuolar sorting receptor
{Vigna mungo}, partial (41%)
Length = 980
Score = 28.9 bits (63), Expect = 6.7
Identities = 13/40 (32%), Positives = 22/40 (54%)
Frame = +2
Query: 398 DAIRRAYPDIQIISNCDGSSRPLDHPADMYDYHIYTNAND 437
D I++A D +++ L HP D +Y ++TN+ND
Sbjct: 644 DRIKKALSDGEMVHINLDWREALPHPDDRVEYELWTNSND 763
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,171,235
Number of Sequences: 36976
Number of extensions: 294039
Number of successful extensions: 1285
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1268
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1278
length of query: 672
length of database: 9,014,727
effective HSP length: 103
effective length of query: 569
effective length of database: 5,206,199
effective search space: 2962327231
effective search space used: 2962327231
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141107.8


