

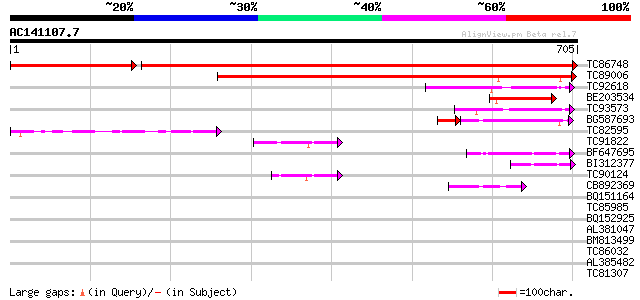
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.7 - phase: 0
(705 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86748 similar to GP|15485584|emb|CAC67503. SET-domain-containi... 1116 0.0
TC89006 weakly similar to GP|20466308|gb|AAM20471.1 unknown prot... 541 e-154
TC92618 weakly similar to PIR|T02416|T02416 probable SET-domain ... 115 8e-26
BE203534 similar to GP|10178033|dbj SET-domain protein-like {Ara... 107 2e-23
TC93573 weakly similar to PIR|T02416|T02416 probable SET-domain ... 86 6e-17
BG587693 weakly similar to GP|17066863|gb Su(VAR)3-9-related pro... 62 2e-14
TC82595 similar to GP|10178033|dbj|BAB11516. SET-domain protein-... 72 7e-13
TC91822 similar to PIR|E96612|E96612 probable transcription fact... 64 3e-10
BF647695 similar to GP|6006866|gb| hypothetical protein {Arabido... 50 3e-06
BI312377 similar to GP|8843772|dbj contains similarity to zinc f... 47 3e-05
TC90124 similar to GP|17529304|gb|AAL38879.1 putative transcript... 45 1e-04
CB892369 weakly similar to GP|18376303|em related to regulatory ... 44 3e-04
BQ151164 42 0.001
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 41 0.002
BQ152925 weakly similar to GP|6448504|emb| Trihydrophobin {Clavi... 37 0.026
AL381047 homologue to PIR|A86193|A86 hypothetical protein [impor... 35 0.076
BM813499 weakly similar to GP|9294325|dbj| gene_id:K24M9.13~unkn... 35 0.13
TC86032 similar to PIR|T07612|T07612 cellulase (EC 3.2.1.4) Cel3... 34 0.22
AL385482 similar to GP|5106924|gb|A putative cell wall protein {... 34 0.22
TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown prot... 33 0.37
>TC86748 similar to GP|15485584|emb|CAC67503. SET-domain-containing protein
{Nicotiana tabacum}, partial (61%)
Length = 2742
Score = 1116 bits (2887), Expect(2) = 0.0
Identities = 542/542 (100%), Positives = 542/542 (100%)
Frame = +1
Query: 164 ATDGSGVAAVDVDLDAVAHDILQSINPMVFDVINHPDGSRDSVTYTLMIYEVLRRKLGQI 223
ATDGSGVAAVDVDLDAVAHDILQSINPMVFDVINHPDGSRDSVTYTLMIYEVLRRKLGQI
Sbjct: 712 ATDGSGVAAVDVDLDAVAHDILQSINPMVFDVINHPDGSRDSVTYTLMIYEVLRRKLGQI 891
Query: 224 EESTKDLHTGAKRPDLKAGNVMMTKGVRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHS 283
EESTKDLHTGAKRPDLKAGNVMMTKGVRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHS
Sbjct: 892 EESTKDLHTGAKRPDLKAGNVMMTKGVRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHS 1071
Query: 284 PSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDTGDGDVLIYSGQGGVNREKGASDQKLE 343
PSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDTGDGDVLIYSGQGGVNREKGASDQKLE
Sbjct: 1072 PSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDTGDGDVLIYSGQGGVNREKGASDQKLE 1251
Query: 344 RGNLALEKSMHRGNDVRVIRGLKDVMHPSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKL 403
RGNLALEKSMHRGNDVRVIRGLKDVMHPSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKL
Sbjct: 1252 RGNLALEKSMHRGNDVRVIRGLKDVMHPSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKL 1431
Query: 404 ARVRGQPEAYTIWKSIQQWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTY 463
ARVRGQPEAYTIWKSIQQWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTY
Sbjct: 1432 ARVRGQPEAYTIWKSIQQWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTY 1611
Query: 464 IPTLKNLRGVAPVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADLKSVIHEC 523
IPTLKNLRGVAPVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADLKSVIHEC
Sbjct: 1612 IPTLKNLRGVAPVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADLKSVIHEC 1791
Query: 524 GPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARA 583
GPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARA
Sbjct: 1792 GPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARA 1971
Query: 584 EMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNV 643
EMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNV
Sbjct: 1972 EMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNV 2151
Query: 644 LWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKNCLCGSVKCRGY 703
LWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKNCLCGSVKCRGY
Sbjct: 2152 LWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKNCLCGSVKCRGY 2331
Query: 704 FC 705
FC
Sbjct: 2332 FC 2337
Score = 302 bits (773), Expect(2) = 0.0
Identities = 149/157 (94%), Positives = 151/157 (95%)
Frame = +2
Query: 1 MDHNLGQESVPADKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAG 60
MDHNLGQESVPADKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAG
Sbjct: 221 MDHNLGQESVPADKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAG 400
Query: 61 VAPFYPFFVSPESQRLSEQHAPNPTPQRATPISAAVPINSFKTPTAATNGDVGSSRRKSR 120
VAPFYPFFVSPESQRLSEQHAPNPTPQRATPISAAVPINSFKTPTAATNGDVGSSRRKSR
Sbjct: 401 VAPFYPFFVSPESQRLSEQHAPNPTPQRATPISAAVPINSFKTPTAATNGDVGSSRRKSR 580
Query: 121 TRRGQLTEEEGYDNTEVIDVDAETGGGSSKRKKRAKG 157
TRRGQLTEEEGYDNTEVIDVDAETGG K ++ KG
Sbjct: 581 TRRGQLTEEEGYDNTEVIDVDAETGGWEFKAQEEGKG 691
Score = 40.8 bits (94), Expect = 0.002
Identities = 19/19 (100%), Positives = 19/19 (100%)
Frame = +3
Query: 145 GGGSSKRKKRAKGRRASGA 163
GGGSSKRKKRAKGRRASGA
Sbjct: 654 GGGSSKRKKRAKGRRASGA 710
>TC89006 weakly similar to GP|20466308|gb|AAM20471.1 unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 1715
Score = 541 bits (1394), Expect = e-154
Identities = 264/467 (56%), Positives = 334/467 (70%), Gaps = 21/467 (4%)
Frame = +3
Query: 259 GIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDTG 318
G VPGVEIGDIFFFR EMC+VGLH+ SM GID L + + EE LAVSIVSSG Y+D+
Sbjct: 3 GSVPGVEIGDIFFFRMEMCVVGLHAQSMGGIDALHIQGDRGEETLAVSIVSSGEYDDEAD 182
Query: 319 DGDVLIYSGQGGV--NREKGASDQKLERGNLALEKSMHRGNDVRVIRGLKDVMHPSGKVY 376
DGDV+IY+GQGG ++K SDQKL +GNLAL++S N++RVIRG+KD ++P K Y
Sbjct: 183 DGDVIIYTGQGGNFNKKDKHVSDQKLHKGNLALDRSSRTHNEIRVIRGIKDAVNPGAKTY 362
Query: 377 VYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYTIWKSIQQWTDKAAPRTGVILPD 436
VYDG+YKIQDSWVEKAK G +FKYKL RV GQP A+ +WKS+Q+W +TG+IL D
Sbjct: 363 VYDGLYKIQDSWVEKAKGGGGLFKYKLIRVPGQPSAFAVWKSVQKWKAGFPAKTGLILAD 542
Query: 437 LTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSFGCSCIG--GCQPGNRN 494
L+SGAE +PV LVN+VDN K PA+FTY +L++ + + ++ S CSC G C PG+ +
Sbjct: 543 LSSGAESLPVSLVNEVDNVKSPAFFTYFHSLRHPKSFSLMQPSHSCSCSGKKACVPGDLD 722
Query: 495 CPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSNK 554
C CI++N G PY G++A+ K ++HECGP+CQC P C+NR+SQ GLK ++EVF+T +K
Sbjct: 723 CSCIRRNEGDFPYIINGVLANRKPLVHECGPTCQCFPNCKNRVSQTGLKHQMEVFKTKDK 902
Query: 555 GWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENE-DEYIFDSTRIYQQLE------- 606
GWGLRSWD IRAG FICEYAGEVID AR L E + DEY+FD+TRIY+ +
Sbjct: 903 GWGLRSWDPIRAGAFICEYAGEVIDKARLSQLVQEGDTDEYVFDTTRIYESFKWNYEPKL 1082
Query: 607 ----VFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAF 662
+ + E +P PL I AKN GNVARFMNHSCSPNV W+P++ E N+ LH+AF
Sbjct: 1083LEEAITNESSEDYALPHPLIINAKNVGNVARFMNHSCSPNVFWQPVLYEENNQSFLHVAF 1262
Query: 663 FAIRHIPPMMELTYDYGINLP-----LQAGQRKKNCLCGSVKCRGYF 704
FA+RHIPPM ELTYDYG + A + +K CLCGS CRG F
Sbjct: 1263FALRHIPPMHELTYDYGSDRSDHTEGSSARKGRKKCLCGSSNCRGSF 1403
>TC92618 weakly similar to PIR|T02416|T02416 probable SET-domain
transcription regulator At2g23750 [imported] -
Arabidopsis thaliana, partial (77%)
Length = 781
Score = 115 bits (287), Expect = 8e-26
Identities = 69/190 (36%), Positives = 103/190 (53%), Gaps = 5/190 (2%)
Frame = +3
Query: 518 SVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEV 577
S++ EC C C TC NRI Q G++ +LEVF T KG+G+R+ +AI GTF+CEY GEV
Sbjct: 3 SLVFECNDKCGCNKTCPNRILQNGVRVKLEVFMTEKKGFGVRAGEAILRGTFVCEYIGEV 182
Query: 578 IDNARA-EMLGAENEDEYIFD----STRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVA 632
++ A G++ Y D + + +E P + I + GNV+
Sbjct: 183 LEQQEAHNRRGSKENCSYFLDIDARANHTSRLVEGHPRYV----------IDSTTYGNVS 332
Query: 633 RFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKN 692
RF+N+SCSPN++ ++ E + HI +A R I ELT++Y P+ + +
Sbjct: 333 RFINNSCSPNLVDYKVLVEATDCKHAHIGLYASRDIALGEELTFNYDYE-PVPG---EGD 500
Query: 693 CLCGSVKCRG 702
CLCGS+KC G
Sbjct: 501 CLCGSLKC*G 530
>BE203534 similar to GP|10178033|dbj SET-domain protein-like {Arabidopsis
thaliana}, partial (12%)
Length = 294
Score = 107 bits (267), Expect = 2e-23
Identities = 53/95 (55%), Positives = 67/95 (69%), Gaps = 11/95 (11%)
Frame = +1
Query: 597 DSTRIYQQ---------LEVFPANI--EAPKIPSPLYITAKNEGNVARFMNHSCSPNVLW 645
D++RIY+ LE +N+ E IPSPL I+A+N GN+ARFMNHSCSPNV W
Sbjct: 1 DTSRIYEPFKWNYEPSLLEDVSSNVCSEDYTIPSPLIISARNVGNIARFMNHSCSPNVFW 180
Query: 646 RPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGI 680
+P++ N+ +HIAFFA+RHIPPM ELTYDYGI
Sbjct: 181 QPVLYAENNQSFIHIAFFALRHIPPMAELTYDYGI 285
>TC93573 weakly similar to PIR|T02416|T02416 probable SET-domain
transcription regulator At2g23750 [imported] -
Arabidopsis thaliana, partial (58%)
Length = 908
Score = 85.5 bits (210), Expect = 6e-17
Identities = 54/153 (35%), Positives = 77/153 (50%), Gaps = 4/153 (2%)
Frame = +3
Query: 554 KGWGLRSWDAIRAGTFICEYAGEVID----NARAEMLGAENEDEYIFDSTRIYQQLEVFP 609
KG G+R+ +AI GTF+CEY GEV+D + R + G N + + R+ +
Sbjct: 27 KGMGVRAGEAILRGTFVCEYIGEVLDVQEAHNRRKRYGTGNCSYFYDINARVNDMSRMIE 206
Query: 610 ANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIP 669
+ I A GNV+RF+NHSCSPN++ ++ E+ + HI F+A + I
Sbjct: 207 EKAQ-------YVIDASKNGNVSRFINHSCSPNLVSHQVLVESMDCERSHIGFYASQDIA 365
Query: 670 PMMELTYDYGINLPLQAGQRKKNCLCGSVKCRG 702
ELTY + L G CLC S KCRG
Sbjct: 366 LGEELTYGFQYELVPGEG---SPCLCESSKCRG 455
>BG587693 weakly similar to GP|17066863|gb Su(VAR)3-9-related protein 4
{Arabidopsis thaliana}, partial (29%)
Length = 688
Score = 61.6 bits (148), Expect(2) = 2e-14
Identities = 47/147 (31%), Positives = 67/147 (44%), Gaps = 6/147 (4%)
Frame = +3
Query: 561 WDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSP 620
W + G F+CE+AGE++ E + + Y L + K
Sbjct: 126 WRNLPKGAFVCEFAGEILTIKELH----ERNIKCAENGKSTYPVLLDADWDSTFVKDEEA 293
Query: 621 LYITAKNEGNVARFMNHSCSP-NVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYG 679
L + A + GN+ARF+NH CS N++ PI E + H A F R+I ELT+DYG
Sbjct: 294 LCLDAASFGNIARFINHRCSDANLVEIPIQIECPDRYYYHFALFTTRNIASHEELTWDYG 473
Query: 680 INL-----PLQAGQRKKNCLCGSVKCR 701
I+ P++ Q C CGS CR
Sbjct: 474 IDFDDHDQPVKLFQ----CKCGSKFCR 542
Score = 35.8 bits (81), Expect(2) = 2e-14
Identities = 16/29 (55%), Positives = 21/29 (72%), Gaps = 1/29 (3%)
Frame = +2
Query: 533 CRNRISQAGLKFRLEVFRTSN-KGWGLRS 560
C NR+ Q G+ + L+VF TS KGWGLR+
Sbjct: 38 CGNRVIQRGITYNLQVFFTSEGKGWGLRT 124
>TC82595 similar to GP|10178033|dbj|BAB11516. SET-domain protein-like
{Arabidopsis thaliana}, partial (7%)
Length = 812
Score = 72.0 bits (175), Expect = 7e-13
Identities = 70/268 (26%), Positives = 114/268 (42%), Gaps = 5/268 (1%)
Frame = +3
Query: 1 MDHNLGQESVPA----DKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGP 56
M+ LGQ SVP DK ++L++KP+R+L+PVF S NPQG +G
Sbjct: 210 MEEGLGQHSVPPPGSIDKYKILDIKPIRSLIPVF--------SKNPQG-------QSSGQ 344
Query: 57 FPAGVAPFYPFFVSPESQRLSEQHAPNPTPQRATPISAAVPINSFKTPTAATNGDVGSSR 116
+P+G +PF+PF H + T + + P+ +F++P
Sbjct: 345 YPSGFSPFFPF---------GGPHDSSTTGAKPRRTAMPTPLQAFRSPFG---------- 467
Query: 117 RKSRTRRGQLTEEEGYDNTEVIDVDAETGGGSSKRKKRAKGRRASGAATDGSGVAAVDVD 176
EE+ DN + + + S R K K + + D SG
Sbjct: 468 ----------EEEDLNDNDDF--SNKRSAASQSTRVKLKKHKVYNDVHVDLSG------- 590
Query: 177 LDAVAHDILQSINPMVFDVINHPDGSRDSVTYTLMIYEVLRRKLGQIEESTKDLHTGAKR 236
L I+P D +G+R+ V LM ++ LRR+L Q+ ++ K+L+TG +
Sbjct: 591 --------LVGISPGQRD-----NGNREVVNTVLMTFDALRRRLSQLVDA-KELNTGFDQ 728
Query: 237 P-DLKAGNVMMTKGVRSNSKKRIGIVPG 263
K+ + + + KR+G VPG
Sbjct: 729 TYXFKSWQYLYDQRNSNKPTKRVGSVPG 812
>TC91822 similar to PIR|E96612|E96612 probable transcription factor
F12K22.14 [imported] - Arabidopsis thaliana, partial
(22%)
Length = 761
Score = 63.5 bits (153), Expect = 3e-10
Identities = 41/114 (35%), Positives = 55/114 (47%), Gaps = 3/114 (2%)
Frame = +1
Query: 304 AVSIVSSGGYEDDTGDGDVLIYSGQGGVNREKGASDQKLERGNLALEKSMHRGNDVRVIR 363
A S+V SGGY D G+ Y+G GG N+ D + N AL S +G VRV+R
Sbjct: 1 AQSVVLSGGYTQDEDHGEWFTYTGSGGRNQ---FLDHQFNNTNEALRLSCRKGYPVRVVR 171
Query: 364 GLKDVMH---PSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYT 414
K+ P V YDG+Y+I W E K+G V +Y R +P +T
Sbjct: 172 SHKEKQSSYAPEAGVR-YDGVYRIDICWSEFGKNGEKVCRYLFVRCDNEPAPWT 330
>BF647695 similar to GP|6006866|gb| hypothetical protein {Arabidopsis
thaliana}, partial (29%)
Length = 460
Score = 50.1 bits (118), Expect = 3e-06
Identities = 41/134 (30%), Positives = 61/134 (44%)
Frame = +3
Query: 569 FICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNE 628
F+ +YAGE++ A+ ++ DE + R L V ++ + K L I A
Sbjct: 6 FLFQYAGELLTTTEAQRR-QQHYDE-LASRGRFSSALLVVREHLPSGKACLRLNIDATRI 179
Query: 629 GNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQ 688
GNVARF+NHSC L +VR + + FFA + I EL + YG ++
Sbjct: 180 GNVARFVNHSCDGGNLSTKLVR-STGALFPRLCFFASKDIQKDEELAFSYG---EIRKRS 347
Query: 689 RKKNCLCGSVKCRG 702
+ C C S C G
Sbjct: 348 NGRLCHCNSPSCLG 389
>BI312377 similar to GP|8843772|dbj contains similarity to zinc finger
protein~gene_id:MYN8.4 {Arabidopsis thaliana}, partial
(7%)
Length = 583
Score = 46.6 bits (109), Expect = 3e-05
Identities = 26/81 (32%), Positives = 43/81 (52%)
Frame = +2
Query: 623 ITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINL 682
+ A ++GN+AR +NHSC PN R + + + + I A ++ ELTYDY +
Sbjct: 2 VDATDKGNIARLINHSCMPNCYARIM---SVGDDESRIVLIAKTNVSAGDELTYDYLFD- 169
Query: 683 PLQAGQRKKNCLCGSVKCRGY 703
P + + K C+C + CR +
Sbjct: 170 PDEPDEFKVPCMCKAPNCRKF 232
>TC90124 similar to GP|17529304|gb|AAL38879.1 putative transcription factor
{Arabidopsis thaliana}, partial (34%)
Length = 1315
Score = 44.7 bits (104), Expect = 1e-04
Identities = 30/92 (32%), Positives = 44/92 (47%), Gaps = 3/92 (3%)
Frame = +2
Query: 326 SGQGGVNREKGASDQKLERGNLALEKSMHRGNDVRVIRGLKD---VMHPSGKVYVYDGIY 382
SG N+ + + DQ+ E N AL S +G VRV+R K+ P V YDG+Y
Sbjct: 20 SGNKRTNKNQ-SFDQQFENMNEALRLSCRKGYPVRVVRSHKEKRSAYAPEAGVR-YDGVY 193
Query: 383 KIQDSWVEKAKSGFNVFKYKLARVRGQPEAYT 414
+I+ W + G V +Y R +P +T
Sbjct: 194 RIEKCWRKIGIQGHKVCRYLFVRCDNEPAPWT 289
>CB892369 weakly similar to GP|18376303|em related to regulatory protein SET1
{Neurospora crassa}, partial (2%)
Length = 740
Score = 43.5 bits (101), Expect = 3e-04
Identities = 31/97 (31%), Positives = 44/97 (44%)
Frame = +2
Query: 546 LEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQL 605
L V+++ G GL + I G + EY GE++ A+ + E EYI Y+
Sbjct: 458 LVVYKSGIHGLGLYTSQCIYRGRMVVEYVGEIVGQRVAD----KREIEYISGRKLQYKSA 625
Query: 606 EVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPN 642
F I I A +G +ARF+NHSC PN
Sbjct: 626 CYFF*------IDKEHIIDATRKGGIARFVNHSCLPN 718
>BQ151164
Length = 772
Score = 41.6 bits (96), Expect = 0.001
Identities = 24/64 (37%), Positives = 38/64 (58%), Gaps = 1/64 (1%)
Frame = +1
Query: 227 TKDLHTGAKRPDLKAG-NVMMTKGVRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHSPS 285
TK+ +T + + G + +TK VRS+ RIG VP ++ +I FF +C+ G+H+ S
Sbjct: 193 TKESNTDSINLTVTKGIHTYLTKRVRSDPPVRIGSVPLFQMENIVFFLIALCVGGMHALS 372
Query: 286 MAGI 289
M GI
Sbjct: 373 MEGI 384
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 40.8 bits (94), Expect = 0.002
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 9/103 (8%)
Frame = +3
Query: 32 SPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAP------FYPFFVSPESQRLSEQHAPNPT 85
SP + +S+P AVSPA P P +P P P+ +S AP P
Sbjct: 225 SPKSSPPASSPTAATVTPAVSPAAPVPVAKSPAASSPVVAPVSTPPKPAPVSSPPAPVPV 404
Query: 86 PQRATPISAAVPINSFKTPTAATNGDV---GSSRRKSRTRRGQ 125
TP+ + P + TP + +V S+ K +T++G+
Sbjct: 405 SSPPTPVPVSSPPTA-STPAVTPSAEVPAAAPSKSKKKTKKGK 530
>BQ152925 weakly similar to GP|6448504|emb| Trihydrophobin {Claviceps
fusiformis}, partial (13%)
Length = 614
Score = 37.0 bits (84), Expect = 0.026
Identities = 25/81 (30%), Positives = 34/81 (41%)
Frame = +2
Query: 28 PVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPESQRLSEQHAPNPTPQ 87
P P+P P + + P V +P P P+G +P +PF P LS P+ P
Sbjct: 281 PTIPNPFQPPTPT------PLVPNNPFLPPPSGSSPLFPF---PSVPGLSPSXPPSSPPG 433
Query: 88 RATPISAAVPINSFKTPTAAT 108
A P P TP A+T
Sbjct: 434 LAFPFPPLFPPPGSGTPPAST 496
>AL381047 homologue to PIR|A86193|A86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (5%)
Length = 490
Score = 35.4 bits (80), Expect = 0.076
Identities = 20/46 (43%), Positives = 23/46 (49%)
Frame = +3
Query: 657 DLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKNCLCGSVKCRG 702
D HI FA R I ELTYDY + + +C CG KCRG
Sbjct: 27 DEHIIIFAKRDIKQWEELTYDY----RFFSIDERLSCYCGFPKCRG 152
>BM813499 weakly similar to GP|9294325|dbj| gene_id:K24M9.13~unknown protein
{Arabidopsis thaliana}, partial (11%)
Length = 709
Score = 34.7 bits (78), Expect = 0.13
Identities = 39/132 (29%), Positives = 60/132 (44%), Gaps = 11/132 (8%)
Frame = +2
Query: 40 SNPQGGAPFVAVSPAGPFPA-GVAPFYPFFVSPESQRLSEQHAPNPT-------PQRAT- 90
S +G A V+++ P PA G+ + P F S E +S AP PT P+ A
Sbjct: 146 SKIEGSANPVSMTFIKPDPAIGLKQYDPLFDSMEPMNISANGAP-PTFSPSIKIPKNAVE 322
Query: 91 --PISAAVPINSFKTPTAATNGDVGSSRRKSRTRRGQLTEEEGYDNTEVIDVDAETGGGS 148
P+ + + N + TN V + S++ +TEE N+ + D+D G
Sbjct: 323 IPPLLSNIGQNCDDSLKKETNKMVAEEKPISQSENN-ITEE----NSPMGDMDQNDGPDE 487
Query: 149 SKRKKRAKGRRA 160
+K+ K AKG RA
Sbjct: 488 AKKTKDAKGSRA 523
>TC86032 similar to PIR|T07612|T07612 cellulase (EC 3.2.1.4) Cel3
membrane-anchored - tomato, complete
Length = 2536
Score = 33.9 bits (76), Expect = 0.22
Identities = 18/49 (36%), Positives = 26/49 (52%)
Frame = +2
Query: 6 GQESVPADKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPA 54
G +S+P DK+ + + P P+FP+P P + P G FV SPA
Sbjct: 1970 GDKSIPIDKNTLFSAVP-----PMFPTPPPPPAPWKP*GVMLFVIFSPA 2101
>AL385482 similar to GP|5106924|gb|A putative cell wall protein {Medicago
truncatula}, partial (42%)
Length = 402
Score = 33.9 bits (76), Expect = 0.22
Identities = 31/96 (32%), Positives = 41/96 (42%), Gaps = 9/96 (9%)
Frame = +2
Query: 29 VFPSPSN---PSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPESQRLSEQHAPNPT 85
V P P+ P + P GGAP +PAGP P G AP +P ++ AP P
Sbjct: 56 VAPPPAGGAPPPGGAPPAGGAPPPGGAPAGPPPEGAAP------TP-----AKTAAPTPG 202
Query: 86 PQRATPISAAVPINSF--KTPTAAT----NGDVGSS 115
+P++ A S K+PT DVG S
Sbjct: 203 GATGSPVAPAGASGSAAPKSPTTGAGVNLKADVGVS 310
>TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 1171
Score = 33.1 bits (74), Expect = 0.37
Identities = 24/67 (35%), Positives = 35/67 (51%), Gaps = 6/67 (8%)
Frame = +2
Query: 31 PSPSNPSSSSNPQGGAPFVAVSPAGPFPA----GVAPFYPFFVSP--ESQRLSEQHAPNP 84
P PS+P+SSS+P +P + S A P P+ ++P F SP S +S H+P
Sbjct: 518 PQPSSPTSSSSP---SPSPSPSSASPSPSLKSFALSPPSSFLHSPFSSSTSVSHGHSPPS 688
Query: 85 TPQRATP 91
+P TP
Sbjct: 689 SP*WKTP 709
Score = 28.9 bits (63), Expect = 7.1
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 2/61 (3%)
Frame = +2
Query: 33 PSNPSSSSNPQGGAP--FVAVSPAGPFPAGVAPFYPFFVSPESQRLSEQHAPNPTPQRAT 90
P +P++SS P F SP PF+ P S S +P+P+P A+
Sbjct: 419 PPSPTASSTASSAVPSNFSLQSPLSS-----PPFFLSLPQPSSPTSSSSPSPSPSPSSAS 583
Query: 91 P 91
P
Sbjct: 584 P 586
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,480,604
Number of Sequences: 36976
Number of extensions: 331203
Number of successful extensions: 2218
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 2124
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2194
length of query: 705
length of database: 9,014,727
effective HSP length: 103
effective length of query: 602
effective length of database: 5,206,199
effective search space: 3134131798
effective search space used: 3134131798
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141107.7


