

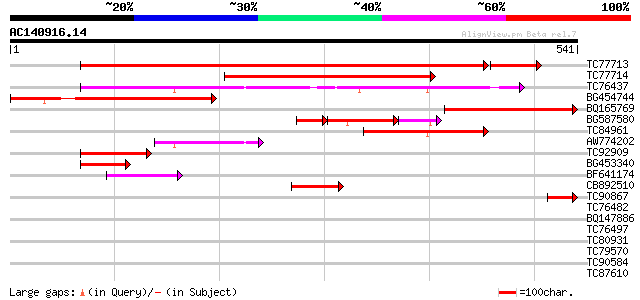
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.14 - phase: 0 /pseudo
(541 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77713 similar to PIR|S62667|S62667 Nramp1 protein - rice, part... 654 0.0
TC77714 similar to PIR|S62667|S62667 Nramp1 protein - rice, part... 384 e-107
TC76437 similar to PIR|T00517|T00517 probable metal ion transpor... 301 5e-82
BG454744 similar to PIR|S62667|S62 Nramp1 protein - rice, partia... 291 6e-79
BQ165769 weakly similar to PIR|S62667|S626 Nramp1 protein - rice... 244 8e-65
BG587580 similar to GP|12666983|emb putative metal transporter {... 102 2e-36
TC84961 similar to PIR|B96513|B96513 stress response protein Nra... 100 2e-21
AW774202 similar to PIR|T00517|T00 probable metal ion transporte... 84 1e-16
TC92909 homologue to PIR|B96513|B96513 stress response protein N... 82 5e-16
BG453340 homologue to PIR|T00517|T005 probable metal ion transpo... 59 4e-09
BF641174 weakly similar to PIR|T48349|T48 EIN2 protein - Arabido... 55 5e-08
CB892510 similar to GP|21554325|gb| metal ion transporter {Arabi... 52 8e-07
TC90867 weakly similar to GP|18176361|gb|AAL60030.1 putative Rab... 46 3e-05
TC76482 similar to PIR|T00517|T00517 probable metal ion transpor... 37 0.015
BQ147886 weakly similar to GP|7242813|emb germin-like protein {P... 35 0.096
TC76497 homologue to SP|P06411|PSBB_TOBAC Photosystem II P680 ch... 33 0.28
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 32 0.47
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 32 0.47
TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Ar... 31 1.1
TC87610 similar to GP|9294102|dbj|BAB01954.1 contains similarity... 31 1.4
>TC77713 similar to PIR|S62667|S62667 Nramp1 protein - rice, partial (84%)
Length = 1575
Score = 654 bits (1688), Expect(2) = 0.0
Identities = 328/390 (84%), Positives = 361/390 (92%)
Frame = +3
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+ETDLQAGANHGYELLW++LIGLIFA IIQSLAANLGV TGKHLSE+C+ EYP FVKYCL
Sbjct: 249 METDLQAGANHGYELLWIILIGLIFACIIQSLAANLGVCTGKHLSEICKAEYPWFVKYCL 428
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
W+LAE+AVIAADIPEVIGTAFALNILF++P+W GVLLTG STLL L LQRFGVRKLELLI
Sbjct: 429 WILAELAVIAADIPEVIGTAFALNILFHVPVWGGVLLTGCSTLLFLGLQRFGVRKLELLI 608
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
+ILVFVMA CFF E+SYV PPA GV++GMFVPKL G GAV DAIALLGALIMPHNLFLHS
Sbjct: 609 SILVFVMAACFFGELSYVKPPAKGVIEGMFVPKLNGNGAVGDAIALLGALIMPHNLFLHS 788
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
ALVLSRK+P + RGIN+ACRYFL ESGFALFVAFLINVAMISV+GTVC A+D+SG+NV+R
Sbjct: 789 ALVLSRKIPGTKRGINDACRYFLIESGFALFVAFLINVAMISVTGTVCKADDISGENVDR 968
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
CND+TLNSASFLL+NVLGRSSSTIYAIALLASGQSS ITGTYAGQ+IMQGFLD++MK+W
Sbjct: 969 CNDITLNSASFLLQNVLGRSSSTIYAIALLASGQSSAITGTYAGQFIMQGFLDLKMKKWI 1148
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
RNL+TR +AIAPSL V+IIGGSSG+ RLIIIASMILSFELPFALIPLLKFSSS TKMGPH
Sbjct: 1149RNLVTRIVAIAPSLVVSIIGGSSGAGRLIIIASMILSFELPFALIPLLKFSSSRTKMGPH 1328
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFV 457
KNS+III ISWILG GII INVYYL TAFV
Sbjct: 1329KNSIIIIIISWILGLGIIGINVYYLITAFV 1418
Score = 86.7 bits (213), Expect(2) = 0.0
Identities = 40/49 (81%), Positives = 45/49 (91%)
Frame = +1
Query: 459 WIIHSSLPKVANVFIGIIVFPLMAIYIASVIYLTFRKDTVEMFIETKND 507
WIIH+SLPKVANVFIG+IVFPLMA+YI SVIYLTFRKDTV F+E K+D
Sbjct: 1423 WIIHNSLPKVANVFIGLIVFPLMALYIVSVIYLTFRKDTVVTFVEVKDD 1569
>TC77714 similar to PIR|S62667|S62667 Nramp1 protein - rice, partial (38%)
Length = 604
Score = 384 bits (986), Expect = e-107
Identities = 200/201 (99%), Positives = 200/201 (99%)
Frame = +1
Query: 206 NPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSALVLSRKVPKSVRGINEA 265
NPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSALVLSRKVPKSVRGINEA
Sbjct: 1 NPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSALVLSRKVPKSVRGINEA 180
Query: 266 CRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERCNDLTLNSASFLLKNVLG 325
CRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERCNDLTLNSASFLLKNVLG
Sbjct: 181 CRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERCNDLTLNSASFLLKNVLG 360
Query: 326 RSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLAVAI 385
RSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLA AI
Sbjct: 361 RSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLAGAI 540
Query: 386 IGGSSGSSRLIIIASMILSFE 406
IGGSSGSSRLIIIASMILSFE
Sbjct: 541 IGGSSGSSRLIIIASMILSFE 603
>TC76437 similar to PIR|T00517|T00517 probable metal ion transporter (NRAMP)
[imported] - Arabidopsis thaliana, partial (92%)
Length = 1694
Score = 301 bits (770), Expect = 5e-82
Identities = 179/431 (41%), Positives = 255/431 (58%), Gaps = 7/431 (1%)
Frame = +3
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E DLQ+GA GY LLWL++ L+IQ L+A LGV TG+HL+E+C EYP + + L
Sbjct: 177 LEGDLQSGAIAGYSLLWLLMWATAMGLLIQLLSARLGVATGRHLAELCRDEYPTWARIVL 356
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
WL+ EVA+I +DI EVIG+A A+ IL N +PLWAGV++T + L L+ +GVRKLE
Sbjct: 357 WLMTEVALIGSDIQEVIGSAIAIRILSNGLVPLWAGVVITALDCFIFLFLENYGVRKLEA 536
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
+L+ +MA F P VL G+ VPKL+ + A+ ++G +IMPHN+FL
Sbjct: 537 FFAVLIAIMALSFAWMFGEAKPDGVDVLVGILVPKLSSR-TIQQAVGVVGCIIMPHNVFL 713
Query: 246 HSALVLSRKV-PKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SR+V PK + EA Y+ ES AL V+F+IN+ + T A G
Sbjct: 714 HSALVQSRQVDPKKKGRVQEALNYYSIESTIALVVSFIINIFV-----TTVFAKGFYGTE 878
Query: 305 VERCNDLTLNSASFLLKNVLGRSSSTIY--AIALLASGQSSTITGTYAGQYIMQGFLDIR 362
+ L +N+ +L + G +Y I LLA+GQSSTITGTYAGQ+IM GFL+++
Sbjct: 879 IANRIGL-VNAGQYLQEKYGGGVFPILYIWGIGLLAAGQSSTITGTYAGQFIMGGFLNLK 1055
Query: 363 MKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLII--IASMILSFELPFALIPLLKFSSS 420
+K+W R L+TR AI P++ VA+I SS S ++ +++ S ++PFALIPLL S
Sbjct: 1056LKKWMRALITRSFAIVPTMIVALIFDSSEESLDVLNEWLNVLQSVQIPFALIPLLCLVSK 1235
Query: 421 STKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPL 480
MG K ++ T+SW++ +I IN Y L F +V+ +V +
Sbjct: 1236ERIMGTFKIGSVLKTVSWLVAALVIVINGYLLLEFFSS--------EVSGAVFTTVVCAI 1391
Query: 481 MAIYIASVIYL 491
A Y+A +IYL
Sbjct: 1392TAAYVAFIIYL 1424
>BG454744 similar to PIR|S62667|S62 Nramp1 protein - rice, partial (29%)
Length = 665
Score = 291 bits (744), Expect = 6e-79
Identities = 158/199 (79%), Positives = 165/199 (82%), Gaps = 2/199 (1%)
Frame = +3
Query: 1 MANLIQQQQQQQQQQQHQVMETSSNHEEP*S--WKQDYTYVFFFHLFLIHFYVFVFVLID 58
MANLIQQQQQQQQQQQHQVMETSSNHEE W++ Y+ L + +
Sbjct: 108 MANLIQQQQQQQQQQQHQVMETSSNHEETQKSGWRKFLPYIGPGFLVSLAY--------- 260
Query: 59 RLFVISQFIVETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVE 118
+ +ETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVE
Sbjct: 261 ----LDPGNMETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVE 428
Query: 119 YPLFVKYCLWLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRF 178
YPLFVKYCLWLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSL F
Sbjct: 429 YPLFVKYCLWLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLXXF 608
Query: 179 GVRKLELLITILVFVMAGC 197
GVRKLELLITILVFVMAGC
Sbjct: 609 GVRKLELLITILVFVMAGC 665
>BQ165769 weakly similar to PIR|S62667|S626 Nramp1 protein - rice, partial
(16%)
Length = 545
Score = 244 bits (622), Expect = 8e-65
Identities = 124/126 (98%), Positives = 124/126 (98%)
Frame = -1
Query: 416 KFSSSSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGI 475
KFSSSSTKMGPHKNSMIIITIS ILG GIISINVYYLSTAFVKWIIHSSLPKVANVFIGI
Sbjct: 545 KFSSSSTKMGPHKNSMIIITISGILGLGIISINVYYLSTAFVKWIIHSSLPKVANVFIGI 366
Query: 476 IVFPLMAIYIASVIYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDL 535
IVFPLMAIYIASVIYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDL
Sbjct: 365 IVFPLMAIYIASVIYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDL 186
Query: 536 ADIPLP 541
ADIPLP
Sbjct: 185 ADIPLP 168
>BG587580 similar to GP|12666983|emb putative metal transporter {Arabidopsis
thaliana}, partial (17%)
Length = 629
Score = 102 bits (255), Expect(3) = 2e-36
Identities = 58/105 (55%), Positives = 66/105 (62%), Gaps = 37/105 (35%)
Frame = +2
Query: 304 NVERCNDLTLNSASFLLK-------------------------------------NVLGR 326
NV+RCND+TLNSASFLL+ NVLGR
Sbjct: 92 NVDRCNDITLNSASFLLQVPNLVRICNYYGQSMILSLILIKQVFTSHLCNFDVD*NVLGR 271
Query: 327 SSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKRNLM 371
SSSTIYAIALLASGQSS ITGTYAGQ+IMQGFLD++MK+W RNL+
Sbjct: 272 SSSTIYAIALLASGQSSAITGTYAGQFIMQGFLDLKMKKWIRNLV 406
Score = 54.3 bits (129), Expect(3) = 2e-36
Identities = 25/30 (83%), Positives = 29/30 (96%)
Frame = +1
Query: 274 GFALFVAFLINVAMISVSGTVCSANDLSGD 303
GFALFVAFLINVAMISV+GTVC A+D+SG+
Sbjct: 1 GFALFVAFLINVAMISVTGTVCKADDISGE 90
Score = 34.7 bits (78), Expect(3) = 2e-36
Identities = 30/74 (40%), Positives = 33/74 (44%), Gaps = 33/74 (44%)
Frame = +3
Query: 372 TRCIAIAPSLAVAII-GGSSGSSRLIIIAS------------------------------ 400
TR +AIAPSL+ GGSSG+ RLIIIAS
Sbjct: 408 TRIVAIAPSLSCLYHWGGSSGAGRLIIIASGLYIPEFAILSSSAYVYILNIIQIT*LKLF 587
Query: 401 --MILSFELPFALI 412
MI SFE PFALI
Sbjct: 588 LQMIPSFEPPFALI 629
>TC84961 similar to PIR|B96513|B96513 stress response protein Nramp2
19015-21280 [imported] - Arabidopsis thaliana, partial
(34%)
Length = 688
Score = 99.8 bits (247), Expect = 2e-21
Identities = 55/122 (45%), Positives = 79/122 (64%), Gaps = 2/122 (1%)
Frame = +1
Query: 338 ASGQSSTITGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLII 397
A+GQSSTITGTYAGQ+IM GFL++R+K+W R L+TR AI P++ VAI+ S +S ++
Sbjct: 112 AAGQSSTITGTYAGQFIMGGFLNLRLKKWLRALITRSCAIVPTIIVAIVYNRSEASLDVL 291
Query: 398 --IASMILSFELPFALIPLLKFSSSSTKMGPHKNSMIIITISWILGFGIISINVYYLSTA 455
+++ S ++PFALIPLL S MG K ++ ++W + II IN Y L
Sbjct: 292 NEWLNVLQSIQIPFALIPLLTLVSKERIMGSFKVGPVLERVAWTVAALIIVINGYLLVDF 471
Query: 456 FV 457
F+
Sbjct: 472 FL 477
>AW774202 similar to PIR|T00517|T00 probable metal ion transporter (NRAMP)
[imported] - Arabidopsis thaliana, partial (20%)
Length = 564
Score = 84.3 bits (207), Expect = 1e-16
Identities = 46/106 (43%), Positives = 62/106 (58%), Gaps = 2/106 (1%)
Frame = -3
Query: 139 DIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLELLITILVFVMAG 196
DI EVIG+A A+ IL N IPLWAGV++T + L L+ +GVR LE +L+ VMA
Sbjct: 331 DIQEVIGSAIAIRILSNGVIPLWAGVVITALDCFIFLFLENYGVRTLEAFFAVLIGVMAL 152
Query: 197 CFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHN 242
F P +L G+ VPKL+ + A+ ++G LIMPHN
Sbjct: 151 AFAWMFGEAKPSGKELLVGILVPKLSSR-TIQQAVGVVGCLIMPHN 17
>TC92909 homologue to PIR|B96513|B96513 stress response protein Nramp2
19015-21280 [imported] - Arabidopsis thaliana, partial
(18%)
Length = 504
Score = 82.0 bits (201), Expect = 5e-16
Identities = 36/68 (52%), Positives = 52/68 (75%)
Frame = +3
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E DLQAGA GY LLWL++ + L+IQSL+A +GV TG+HL+E+C+ EYP + +Y L
Sbjct: 300 LEGDLQAGAIAGYSLLWLLMWATLMGLLIQSLSARVGVATGRHLAELCKEEYPNWARYVL 479
Query: 128 WLLAEVAV 135
W +AE+A+
Sbjct: 480 WFMAELAL 503
>BG453340 homologue to PIR|T00517|T005 probable metal ion transporter (NRAMP)
[imported] - Arabidopsis thaliana, partial (14%)
Length = 472
Score = 59.3 bits (142), Expect = 4e-09
Identities = 28/48 (58%), Positives = 35/48 (72%)
Frame = +2
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVC 115
+E DLQAGA GY LLWL++ L+IQ L+A LGV TGKHL+E+C
Sbjct: 326 LEGDLQAGAIAGYSLLWLLMWATAMGLLIQLLSARLGVATGKHLAELC 469
>BF641174 weakly similar to PIR|T48349|T48 EIN2 protein - Arabidopsis
thaliana, partial (6%)
Length = 684
Score = 55.5 bits (132), Expect = 5e-08
Identities = 28/73 (38%), Positives = 43/73 (58%)
Frame = +2
Query: 93 ALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCLWLLAEVAVIAADIPEVIGTAFALNI 152
A+ Q L+A +GV TG+HL+++C EY L + AE++VI D+ ++G A LN+
Sbjct: 422 AIFFQYLSARVGVITGRHLAQICSDEYDTLTCLLLGIQAELSVIMLDLNMILGMAQGLNL 601
Query: 153 LFNIPLWAGVLLT 165
+F L V LT
Sbjct: 602 IFGWELXTCVFLT 640
>CB892510 similar to GP|21554325|gb| metal ion transporter {Arabidopsis
thaliana}, partial (7%)
Length = 687
Score = 51.6 bits (122), Expect = 8e-07
Identities = 26/49 (53%), Positives = 34/49 (69%)
Frame = +3
Query: 270 LYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERCNDLTLNSASF 318
+ ES FAL VAFLINV++ISVSG V + +++ + C DL LN ASF
Sbjct: 540 MIESAFALMVAFLINVSVISVSGAVLHSPNINEERSMSCQDLDLNKASF 686
>TC90867 weakly similar to GP|18176361|gb|AAL60030.1 putative Rab
geranylgeranyl transferase {Arabidopsis thaliana},
partial (6%)
Length = 697
Score = 46.2 bits (108), Expect = 3e-05
Identities = 21/28 (75%), Positives = 24/28 (85%)
Frame = +3
Query: 514 VEKGIVDNGQLELSHVPYREDLADIPLP 541
VEKG V++GQLELS+ PYREDLADI P
Sbjct: 9 VEKGFVNDGQLELSYTPYREDLADITRP 92
>TC76482 similar to PIR|T00517|T00517 probable metal ion transporter (NRAMP)
[imported] - Arabidopsis thaliana, partial (8%)
Length = 599
Score = 37.4 bits (85), Expect = 0.015
Identities = 15/41 (36%), Positives = 25/41 (60%)
Frame = +2
Query: 151 NILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLITILV 191
+++ N LWAGV++T + L L+ +GVR LE +L+
Sbjct: 473 HLMRNQSLWAGVVITALDCFIFLFLENYGVRTLEAFFAVLI 595
>BQ147886 weakly similar to GP|7242813|emb germin-like protein {Phaseolus
vulgaris}, partial (73%)
Length = 646
Score = 34.7 bits (78), Expect = 0.096
Identities = 22/46 (47%), Positives = 29/46 (62%), Gaps = 1/46 (2%)
Frame = +2
Query: 34 QDYTYVFFFHLFLIHFYVFVFVLIDRLFVISQFIVETDLQA-GANH 78
+DY+Y F FH FLIH +V V +I FV+ ++T LQA ANH
Sbjct: 5 EDYSYTFPFHRFLIHCFVCVLSMI---FVLRI*RLQTLLQAMPANH 133
>TC76497 homologue to SP|P06411|PSBB_TOBAC Photosystem II P680 chlorophyll A
apoprotein (CP-47 protein). [Common tobacco] {Nicotiana
tabacum}, complete
Length = 4045
Score = 33.1 bits (74), Expect = 0.28
Identities = 13/23 (56%), Positives = 18/23 (77%)
Frame = -1
Query: 41 FFHLFLIHFYVFVFVLIDRLFVI 63
FF L+LI FY F F++IDR F++
Sbjct: 3031 FFCLYLIRFYCFFFIIIDRNFLV 2963
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 32.3 bits (72), Expect = 0.47
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +1
Query: 6 QQQQQQQQQQQHQVMETSSNH 26
QQQ QQQQ QQHQ M++ H
Sbjct: 493 QQQMQQQQMQQHQQMQSQQQH 555
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 32.3 bits (72), Expect = 0.47
Identities = 13/23 (56%), Positives = 17/23 (73%)
Frame = +2
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQQQQQQQQQHQ + + ++
Sbjct: 593 QQQQQQQQQQQHQQQQQQQSQQQ 661
Score = 30.0 bits (66), Expect = 2.4
Identities = 14/30 (46%), Positives = 19/30 (62%)
Frame = +2
Query: 5 IQQQQQQQQQQQHQVMETSSNHEEP*SWKQ 34
+QQQQQQQQQQQ Q + ++ S +Q
Sbjct: 572 MQQQQQQQQQQQQQQQQQHQQQQQQQSQQQ 661
Score = 28.5 bits (62), Expect = 6.8
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = +2
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQQQQQQQQ Q + S ++
Sbjct: 596 QQQQQQQQQQHQQQQQQQSQQQQ 664
Score = 28.5 bits (62), Expect = 6.8
Identities = 12/23 (52%), Positives = 16/23 (69%)
Frame = +2
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQ QQQQQQQ Q + +H++
Sbjct: 617 QQQHQQQQQQQSQQQQQVVDHQQ 685
Score = 28.5 bits (62), Expect = 6.8
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = +2
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQQQQQQQ Q Q + S ++
Sbjct: 599 QQQQQQQQQHQQQQQQQSQQQQQ 667
>TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Arabidopsis
thaliana}, partial (18%)
Length = 686
Score = 31.2 bits (69), Expect = 1.1
Identities = 12/33 (36%), Positives = 21/33 (63%)
Frame = +2
Query: 34 QDYTYVFFFHLFLIHFYVFVFVLIDRLFVISQF 66
+D T++FFF F + ++F+F I R F + +F
Sbjct: 86 KDGTHLFFFVFFFLRCFIFLFFFIFRSFFLQRF 184
>TC87610 similar to GP|9294102|dbj|BAB01954.1 contains similarity to AAA-type
ATPase~gene_id:T20D4.3 {Arabidopsis thaliana}, partial
(53%)
Length = 1765
Score = 30.8 bits (68), Expect = 1.4
Identities = 10/26 (38%), Positives = 20/26 (76%)
Frame = -3
Query: 42 FHLFLIHFYVFVFVLIDRLFVISQFI 67
FH L+HF++ +F+ ++ F++SQF+
Sbjct: 1604 FHTILLHFFL*LFISLNLFFLLSQFL 1527
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.141 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,294,160
Number of Sequences: 36976
Number of extensions: 260487
Number of successful extensions: 3982
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 2920
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3449
length of query: 541
length of database: 9,014,727
effective HSP length: 101
effective length of query: 440
effective length of database: 5,280,151
effective search space: 2323266440
effective search space used: 2323266440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140916.14


