

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.1 - phase: 0
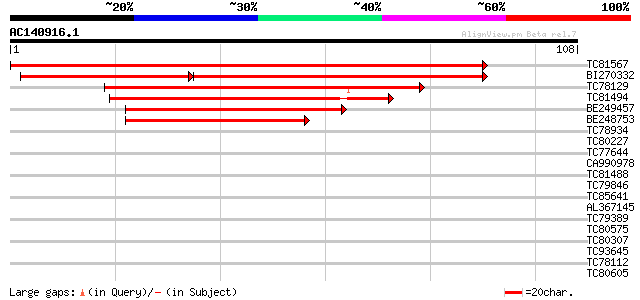
(108 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81567 similar to PIR|T47764|T47764 responce reactor 4 - Arabid... 178 2e-46
BI270332 similar to PIR|T06673|T066 response reactor 2 [imported... 110 4e-40
TC78129 similar to PIR|T47764|T47764 responce reactor 4 - Arabid... 65 4e-12
TC81494 similar to GP|13173408|gb|AAK14395.1 response regulator ... 64 9e-12
BE249457 similar to PIR|T50854|T508 response regulator 7 [import... 59 3e-10
BE248753 homologue to GP|11870065|gb| response regulator 15 {Ara... 51 8e-08
TC78934 similar to GP|14189890|dbj|BAB55874. response regulator ... 32 0.038
TC80227 similar to GP|13537198|dbj|BAB40775. histidine kinase {A... 31 0.084
TC77644 similar to GP|16751567|gb|AAL27697.1 5-enolpyruvylshikim... 30 0.14
CA990978 weakly similar to GP|12002863|gb|A self-pruning interac... 29 0.32
TC81488 GP|17985051|gb|AAL54093.1 EXOPOLYSACCHARIDE PRODUCTION P... 28 0.42
TC79846 similar to GP|8843728|dbj|BAA97276.1 homeodomain transcr... 28 0.55
TC85641 weakly similar to GP|8777295|dbj|BAA96885.1 cytochrome P... 28 0.71
AL367145 weakly similar to GP|8096269|dbj| KED {Nicotiana tabacu... 27 0.93
TC79389 similar to GP|14189890|dbj|BAB55874. response regulator ... 27 0.93
TC80575 weakly similar to GP|12056928|gb|AAG48132.1 putative res... 27 1.6
TC80307 similar to GP|20160990|dbj|BAB89924. putative protein ki... 26 2.1
TC93645 similar to GP|19699053|gb|AAL90894.1 AT5g23390/T32G24_2 ... 26 2.1
TC78112 similar to PIR|C96673|C96673 gamma-tocopherol methyltran... 26 2.1
TC80605 similar to PIR|S56716|S56716 protein kinase SPK-3 (EC 2.... 25 3.5
>TC81567 similar to PIR|T47764|T47764 responce reactor 4 - Arabidopsis
thaliana, partial (27%)
Length = 470
Score = 178 bits (452), Expect = 2e-46
Identities = 91/91 (100%), Positives = 91/91 (100%)
Frame = +3
Query: 1 MELGLDSRKLQLQPQNLKQEHFHVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKY 60
MELGLDSRKLQLQPQNLKQEHFHVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKY
Sbjct: 36 MELGLDSRKLQLQPQNLKQEHFHVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKY 215
Query: 61 LGLNIDEISSTKSSILESSSPSLPQPLQLQE 91
LGLNIDEISSTKSSILESSSPSLPQPLQLQE
Sbjct: 216 LGLNIDEISSTKSSILESSSPSLPQPLQLQE 308
>BI270332 similar to PIR|T06673|T066 response reactor 2 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 556
Score = 110 bits (275), Expect(2) = 4e-40
Identities = 56/56 (100%), Positives = 56/56 (100%)
Frame = +2
Query: 36 LLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKSSILESSSPSLPQPLQLQE 91
LLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKSSILESSSPSLPQPLQLQE
Sbjct: 101 LLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKSSILESSSPSLPQPLQLQE 268
Score = 68.9 bits (167), Expect(2) = 4e-40
Identities = 33/33 (100%), Positives = 33/33 (100%)
Frame = +1
Query: 3 LGLDSRKLQLQPQNLKQEHFHVLAVDDSVIDRM 35
LGLDSRKLQLQPQNLKQEHFHVLAVDDSVIDRM
Sbjct: 1 LGLDSRKLQLQPQNLKQEHFHVLAVDDSVIDRM 99
>TC78129 similar to PIR|T47764|T47764 responce reactor 4 - Arabidopsis
thaliana, partial (62%)
Length = 1307
Score = 65.1 bits (157), Expect = 4e-12
Identities = 34/62 (54%), Positives = 45/62 (71%), Gaps = 1/62 (1%)
Frame = +1
Query: 19 QEHFHVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYLGL-NIDEISSTKSSILE 77
+ FHVLAVDDS+IDR L+ERLL+ SS +VT VDSG KAL++LGL DE + S+
Sbjct: 394 ESQFHVLAVDDSIIDRKLIERLLKTSSYQVTTVDSGSKALEFLGLCENDETNPNTPSVFP 573
Query: 78 SS 79
++
Sbjct: 574 NN 579
>TC81494 similar to GP|13173408|gb|AAK14395.1 response regulator protein
{Dianthus caryophyllus}, partial (53%)
Length = 793
Score = 63.9 bits (154), Expect = 9e-12
Identities = 31/54 (57%), Positives = 45/54 (82%)
Frame = +1
Query: 20 EHFHVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKS 73
+ HVLAVDDS++DR ++ERLL+ S+CKVT VDSG +AL++LGL +DE +++S
Sbjct: 82 QEVHVLAVDDSLVDRKVIERLLKISACKVTAVDSGIRALQFLGL-VDEKETSES 240
>BE249457 similar to PIR|T50854|T508 response regulator 7 [imported] -
Arabidopsis thaliana, partial (21%)
Length = 246
Score = 58.9 bits (141), Expect = 3e-10
Identities = 27/42 (64%), Positives = 37/42 (87%)
Frame = +2
Query: 23 HVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYLGLN 64
HVLAVDDS++DR ++ERLL+ SSCKVT V+S +AL++LGL+
Sbjct: 116 HVLAVDDSLVDRKVIERLLKISSCKVTVVESARRALQFLGLD 241
>BE248753 homologue to GP|11870065|gb| response regulator 15 {Arabidopsis
thaliana}, partial (22%)
Length = 257
Score = 50.8 bits (120), Expect = 8e-08
Identities = 23/35 (65%), Positives = 30/35 (85%)
Frame = +2
Query: 23 HVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKA 57
HVLAVDDS +DR ++ERLL+ +SCKVT V+SG +A
Sbjct: 122 HVLAVDDSFVDRKVIERLLKFTSCKVTVVESGTRA 226
>TC78934 similar to GP|14189890|dbj|BAB55874. response regulator 9 {Zea
mays}, partial (25%)
Length = 1014
Score = 32.0 bits (71), Expect = 0.038
Identities = 20/43 (46%), Positives = 21/43 (48%)
Frame = +2
Query: 24 VLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYLGLNID 66
VLAVDD +LLE LLR VT AL L NID
Sbjct: 68 VLAVDDDPTSLLLLETLLRSCQYHVTTTSEAITALTMLQENID 196
>TC80227 similar to GP|13537198|dbj|BAB40775. histidine kinase {Arabidopsis
thaliana}, partial (22%)
Length = 1693
Score = 30.8 bits (68), Expect = 0.084
Identities = 19/60 (31%), Positives = 27/60 (44%), Gaps = 4/60 (6%)
Frame = +3
Query: 6 DSRKLQLQPQNLKQEHF----HVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYL 61
+ R L QNL H +L VDD+ ++R + L+ V CV SG A+ L
Sbjct: 702 NKRTLNRDLQNLSLSHLLVGRKILIVDDNGVNRAVAAGALKKYGADVVCVSSGKDAISSL 881
>TC77644 similar to GP|16751567|gb|AAL27697.1
5-enolpyruvylshikimate-3-phosphate synthase {Dicliptera
chinensis}, partial (41%)
Length = 1342
Score = 30.0 bits (66), Expect = 0.14
Identities = 20/62 (32%), Positives = 35/62 (56%), Gaps = 6/62 (9%)
Frame = +1
Query: 30 SVIDRMLLERLLRDSSCKV-TCVDSGD-----KALKYLGLNIDEISSTKSSILESSSPSL 83
S+ +R+LL L + + V +DS D +ALK LGL +++ +TK +++E S
Sbjct: 535 SLSNRILLLAALSEGTTVVENLLDSEDIHYMLEALKTLGLRVEDDKTTKQAVVEGSGGLF 714
Query: 84 PQ 85
P+
Sbjct: 715 PE 720
>CA990978 weakly similar to GP|12002863|gb|A self-pruning interacting protein
1 {Lycopersicon esculentum}, partial (24%)
Length = 657
Score = 28.9 bits (63), Expect = 0.32
Identities = 15/46 (32%), Positives = 25/46 (53%), Gaps = 1/46 (2%)
Frame = +2
Query: 49 TCVDSGDKALKYLGLNIDEI-SSTKSSILESSSPSLPQPLQLQEVI 93
TC+ G K K+L + + I K E ++P+P+Q++EVI
Sbjct: 119 TCLQQGQKRHKFLVICEESIIKQEKREQEEEEEANIPKPIQVEEVI 256
>TC81488 GP|17985051|gb|AAL54093.1 EXOPOLYSACCHARIDE PRODUCTION PROTEIN EXOF
PRECURSOR {Brucella melitensis}, partial (3%)
Length = 921
Score = 28.5 bits (62), Expect = 0.42
Identities = 12/31 (38%), Positives = 22/31 (70%)
Frame = +3
Query: 30 SVIDRMLLERLLRDSSCKVTCVDSGDKALKY 60
S I +++ +LRDSSC ++C+ + D A++Y
Sbjct: 513 SRISSLVIAPILRDSSCFISCI-AADNAVRY 602
>TC79846 similar to GP|8843728|dbj|BAA97276.1 homeodomain transcription
factor-like {Arabidopsis thaliana}, partial (42%)
Length = 679
Score = 28.1 bits (61), Expect = 0.55
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 14/88 (15%)
Frame = +2
Query: 1 MELGLDSRKLQLQPQN---------LKQEHFHVLAVDDS-VIDRMLLERLLRDSSCKV-- 48
MELGLD R++ + QN L++E+F + +S ++++ LLE LR+ +
Sbjct: 356 MELGLDPRQVAVWFQNRRARWKNKKLEEEYFSLKKNHESTILEKCLLETKLREQHSEALK 535
Query: 49 --TCVDSGDKALKYLGLNIDEISSTKSS 74
+ +K ++ L + ++S+ SS
Sbjct: 536 LREQLSEAEKEIQRLREPFERVTSSSSS 619
>TC85641 weakly similar to GP|8777295|dbj|BAA96885.1 cytochrome P450-like
{Arabidopsis thaliana}, partial (71%)
Length = 1941
Score = 27.7 bits (60), Expect = 0.71
Identities = 16/45 (35%), Positives = 25/45 (55%)
Frame = -1
Query: 21 HFHVLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYLGLNI 65
+FH+L +D + R L E ++S+C + V S D+ K L NI
Sbjct: 1077 NFHLLLIDFIINVR*LAEIFDKESACDASSVMSSDQKAKNLISNI 943
>AL367145 weakly similar to GP|8096269|dbj| KED {Nicotiana tabacum}, partial
(8%)
Length = 284
Score = 27.3 bits (59), Expect = 0.93
Identities = 15/37 (40%), Positives = 20/37 (53%)
Frame = +2
Query: 69 SSTKSSILESSSPSLPQPLQLQEVIILIFFFLILKSF 105
SS ++L S S P L L +L+FFFL+L F
Sbjct: 158 SSFCFNLLTSLDLSFPS*LSLLSTFVLVFFFLLLFHF 268
>TC79389 similar to GP|14189890|dbj|BAB55874. response regulator 9 {Zea
mays}, partial (33%)
Length = 1532
Score = 27.3 bits (59), Expect = 0.93
Identities = 19/52 (36%), Positives = 23/52 (43%)
Frame = +1
Query: 24 VLAVDDSVIDRMLLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKSSI 75
VLAVDD ++LE LLR VT AL L N D+ S +
Sbjct: 211 VLAVDDDPTCLLVLETLLRRCQYHVTTTSQAITALTMLRENKDKFDLVISDV 366
>TC80575 weakly similar to GP|12056928|gb|AAG48132.1 putative resistance
protein {Glycine max}, partial (5%)
Length = 758
Score = 26.6 bits (57), Expect = 1.6
Identities = 14/46 (30%), Positives = 24/46 (51%)
Frame = -3
Query: 32 IDRMLLERLLRDSSCKVTCVDSGDKALKYLGLNIDEISSTKSSILE 77
+ + ++ L++ SCK G K L L LN+ +ISS ++E
Sbjct: 135 LTQSVISNFLKEGSCK------GGKCLSSLQLNVSKISSLFRRVME 16
>TC80307 similar to GP|20160990|dbj|BAB89924. putative protein kinase {Oryza
sativa (japonica cultivar-group)}, partial (69%)
Length = 1929
Score = 26.2 bits (56), Expect = 2.1
Identities = 11/32 (34%), Positives = 21/32 (65%)
Frame = -3
Query: 64 NIDEISSTKSSILESSSPSLPQPLQLQEVIIL 95
N+D + + + +++ +S+ SLP PL L E + L
Sbjct: 1600 NLDSLQTARLALIGNSTTSLPCPLGLVEALEL 1505
>TC93645 similar to GP|19699053|gb|AAL90894.1 AT5g23390/T32G24_2
{Arabidopsis thaliana}, partial (26%)
Length = 994
Score = 26.2 bits (56), Expect = 2.1
Identities = 10/22 (45%), Positives = 16/22 (72%)
Frame = -2
Query: 86 PLQLQEVIILIFFFLILKSFQL 107
PL LQE +++ FFFL L+ + +
Sbjct: 579 PLLLQETLLIAFFFLHLQKYHV 514
>TC78112 similar to PIR|C96673|C96673 gamma-tocopherol methyltransferase
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1100
Score = 26.2 bits (56), Expect = 2.1
Identities = 15/42 (35%), Positives = 21/42 (49%)
Frame = -2
Query: 58 LKYLGLNIDEISSTKSSILESSSPSLPQPLQLQEVIILIFFF 99
L L + ID ++ SS+L PSL P L + L FF+
Sbjct: 238 LHSLAILIDPLTEVSSSLLLPLPPSLLSPPPLIHAMNLFFFY 113
>TC80605 similar to PIR|S56716|S56716 protein kinase SPK-3 (EC 2.7.1.-) -
soybean, partial (25%)
Length = 533
Score = 25.4 bits (54), Expect = 3.5
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = -2
Query: 69 SSTKSSILESSSPSLPQPL 87
SS+KSS +SSPS P+PL
Sbjct: 202 SSSKSSSPSTSSPSQPKPL 146
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,265,055
Number of Sequences: 36976
Number of extensions: 41047
Number of successful extensions: 343
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 341
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 343
length of query: 108
length of database: 9,014,727
effective HSP length: 84
effective length of query: 24
effective length of database: 5,908,743
effective search space: 141809832
effective search space used: 141809832
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 50 (23.9 bits)
Medicago: description of AC140916.1


