

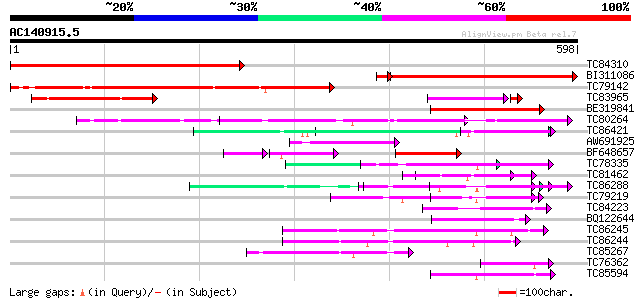
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140915.5 - phase: 0
(598 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84310 similar to GP|18844836|dbj|BAB85306. hypothetical protei... 487 e-138
BI311086 396 e-115
TC79142 266 2e-71
TC83965 weakly similar to GP|21592320|gb|AAM64271.1 unknown {Ara... 168 6e-42
BE319841 weakly similar to GP|21592320|gb| unknown {Arabidopsis ... 124 1e-28
TC80264 similar to GP|21689699|gb|AAM67471.1 putative F-box fami... 71 1e-12
TC86421 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max},... 69 7e-12
AW691925 67 3e-11
BF648657 64 2e-10
TC78335 similar to GP|16604366|gb|AAL24189.1 At1g21410/F24J8_17 ... 63 4e-10
TC81462 similar to GP|21536497|gb|AAM60829.1 F-box protein famil... 60 3e-09
TC86288 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max},... 60 3e-09
TC79219 similar to GP|15810000|gb|AAL06927.1 AT5g23340/MKD15_20 ... 60 3e-09
TC84223 weakly similar to GP|18252175|gb|AAL61920.1 unknown prot... 57 3e-08
BQ122644 56 4e-08
TC86245 weakly similar to GP|9279671|dbj|BAB01228.1 transport in... 54 2e-07
TC86244 weakly similar to PIR|T48087|T48087 transport inhibitor ... 52 8e-07
TC85267 similar to GP|9279671|dbj|BAB01228.1 transport inhibitor... 48 9e-06
TC76362 similar to GP|10716949|gb|AAG21977.1 SKP1 interacting pa... 46 4e-05
TC85594 similar to PIR|D96512|D96512 hypothetical protein F2G19.... 46 4e-05
>TC84310 similar to GP|18844836|dbj|BAB85306. hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (2%)
Length = 799
Score = 487 bits (1254), Expect = e-138
Identities = 247/247 (100%), Positives = 247/247 (100%)
Frame = +3
Query: 1 MKRQRTNSFKLNSCSQQLNSSSSSELDLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVAS 60
MKRQRTNSFKLNSCSQQLNSSSSSELDLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVAS
Sbjct: 57 MKRQRTNSFKLNSCSQQLNSSSSSELDLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVAS 236
Query: 61 KHFLSVTNRHKFCLTILYPALPVLPGLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLK 120
KHFLSVTNRHKFCLTILYPALPVLPGLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLK
Sbjct: 237 KHFLSVTNRHKFCLTILYPALPVLPGLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLK 416
Query: 121 LTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAY 180
LTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAY
Sbjct: 417 LTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAY 596
Query: 181 PSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRC 240
PSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRC
Sbjct: 597 PSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRC 776
Query: 241 EQLTIAG 247
EQLTIAG
Sbjct: 777 EQLTIAG 797
>BI311086
Length = 718
Score = 396 bits (1018), Expect(2) = e-115
Identities = 196/197 (99%), Positives = 197/197 (99%)
Frame = +1
Query: 402 QGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGI 461
+GTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGI
Sbjct: 46 RGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGI 225
Query: 462 RPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQL 521
RPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQL
Sbjct: 226 RPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQL 405
Query: 522 VLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHVPPNFPL 581
VLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHVPPNFPL
Sbjct: 406 VLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHVPPNFPL 585
Query: 582 SDRNRKLFSRHGCLLVM 598
SDRNRKLFSRHGCLLVM
Sbjct: 586 SDRNRKLFSRHGCLLVM 636
Score = 38.5 bits (88), Expect = 0.007
Identities = 43/188 (22%), Positives = 80/188 (41%), Gaps = 22/188 (11%)
Frame = +1
Query: 274 LTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLL 333
+ + +D L++ + + L + + DQ + A L++L LS T GI LL
Sbjct: 58 IPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLL 237
Query: 334 SKSKRIQHLDLQY-------TDF--------------LNDHCVAELSLFLGDLLSLNLGN 372
++I+HL+L T+F ++D + +S LL L L
Sbjct: 238 ESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLR 417
Query: 373 CRLLTVSTFFALITNCPSLTEINMNR-TNIQGTTIPNSLMDRLVNPQFKSLFLASAACLE 431
C +T ++ NC L EIN++ N+Q + + ++ R P + + + L
Sbjct: 418 CDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSR---PSLRKIHVPPNFPLS 588
Query: 432 DQNIIMFA 439
D+N +F+
Sbjct: 589 DRNRKLFS 612
Score = 37.7 bits (86), Expect(2) = e-115
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +3
Query: 388 CPSLTEINMNRTNIQG 403
CPSLTEINMNRTNIQG
Sbjct: 3 CPSLTEINMNRTNIQG 50
>TC79142
Length = 1164
Score = 266 bits (680), Expect = 2e-71
Identities = 169/357 (47%), Positives = 232/357 (64%), Gaps = 15/357 (4%)
Frame = +3
Query: 1 MKRQRTNSFKLNSCSQQLNSSSSSELDLPDDIWERVFR-LLKNNDDDDHRKRY-LKSLSV 58
MKR+R++S K SQQ SS DLP++ WE +FR +LK+ ++ D+ Y L SLS+
Sbjct: 129 MKRKRSSSTK---SSQQAVSS-----DLPNECWELIFRFILKDENNLDYYSLYILNSLSL 284
Query: 59 ASKHFLSVTNRHKFCLTILYPALPVLPGLLQRFTKLTSLDLS-YYYGDLDALLTQISSFP 117
SK FLS+TN + L++ + + ++P + +RFT +TSL + Y DLD +L ++S FP
Sbjct: 285 VSKQFLSITNTLRLSLSVPF-SRGMIP-IFKRFTDITSLKFDPFLYPDLDKILCKVSRFP 458
Query: 118 MLKLTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELD 177
+ KLTSL++S Q +PANGLR FSQNI TLTSL CS++ N+TD+ LIA+ FPLLEEL+
Sbjct: 459 LKKLTSLDISYQCTIPANGLRVFSQNIRTLTSLTCSDIDDFNTTDLFLIAECFPLLEELN 638
Query: 178 LAYPS--KIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEV 235
L +PS K I + + G+EALSLAL+KLRKV L ++ L +LF +CK L++V
Sbjct: 639 LIFPSDWKSIYKNYNNYCDGIEALSLALVKLRKVKLCRFP-MSDKSLFYLFNSCKLLEDV 815
Query: 236 ILLRCEQLTIAGVDLALLQKPTLTSLSITCTVT--------TGLEH--LTSHFIDSLLSL 285
L C+Q+TIAG+ A+ ++PTL SLS C T + H +TS FIDSL+SL
Sbjct: 816 TLFECDQITIAGIASAIRERPTLKSLSF-CNRTYSENSISNDDISHFKVTSDFIDSLVSL 992
Query: 286 KGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHL 342
KGLTSL L ISD+ L SIA E LPL RL L YC +Y+GI L S + +QH+
Sbjct: 993 KGLTSLSLRSLDISDEMLYSIAREGLPLTRLALRYCLD*SYAGIFCLSSSVRGLQHM 1163
Score = 41.2 bits (95), Expect = 0.001
Identities = 44/158 (27%), Positives = 73/158 (45%), Gaps = 19/158 (12%)
Frame = +3
Query: 435 IIMFAALFP--NLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDL-- 490
I+ + FP L L +S I G+R ++ R + +LTC + T DL
Sbjct: 432 ILCKVSRFPLKKLTSLDISYQCTIPANGLRVFSQNIRTLT--SLTCSDIDDFNTT-DLFL 602
Query: 491 -----PDLEVLNLTNTEVDDEALYI-ISNRCPAL----LQLVLLR----CDY-ITDKGVM 535
P LE LNL D +++Y +N C + L LV LR C + ++DK +
Sbjct: 603 IAECFPLLEELNLIFPS-DWKSIYKNYNNYCDGIEALSLALVKLRKVKLCRFPMSDKSLF 779
Query: 536 HVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKI 573
++ N+C L ++ L C + +AS + RP+L+ +
Sbjct: 780 YLFNSCKLLEDVTLFECDQITIAGIASAIRERPTLKSL 893
Score = 34.3 bits (77), Expect = 0.14
Identities = 33/149 (22%), Positives = 67/149 (44%), Gaps = 5/149 (3%)
Frame = +3
Query: 405 TIPNSLMDRLVNPQFKSLFLASAACLEDQN---IIMFAALFPNLQQLHLSCSYNITEEGI 461
TIP + + R+ + ++L + + ++D N + + A FP L++L+L
Sbjct: 498 TIPANGL-RVFSQNIRTLTSLTCSDIDDFNTTDLFLIAECFPLLEELNL----------- 641
Query: 462 RPLLESCRK--IRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALL 519
+ S K ++ N C +++L + L L + L + D++L+ + N C L
Sbjct: 642 --IFPSDWKSIYKNYNNYCDGIEAL--SLALVKLRKVKLCRFPMSDKSLFYLFNSCKLLE 809
Query: 520 QLVLLRCDYITDKGVMHVVNNCTQLREIN 548
+ L CD IT G+ + L+ ++
Sbjct: 810 DVTLFECDQITIAGIASAIRERPTLKSLS 896
>TC83965 weakly similar to GP|21592320|gb|AAM64271.1 unknown {Arabidopsis
thaliana}, partial (6%)
Length = 745
Score = 168 bits (425), Expect = 6e-42
Identities = 91/134 (67%), Positives = 103/134 (75%), Gaps = 1/134 (0%)
Frame = +2
Query: 24 SELDLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTILYPALPV 83
++ DLPDD W +FR L N+DDD+H++RY++SLSV+SKHFLSVTNRHKF LTI LP
Sbjct: 53 NDFDLPDDCWRCIFRFL-NDDDDNHKQRYMESLSVSSKHFLSVTNRHKFSLTISDQTLPS 229
Query: 84 LPGLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLKLTSLNLSNQLILPANGLRAFSQN 143
LP L QRFT LTSLD + GDLD LL QISSFP L LTSLNLSN I+P NGLR FSQN
Sbjct: 230 LPRLFQRFTNLTSLDFKRFCGDLDTLLCQISSFP-LNLTSLNLSNHPIIPQNGLRVFSQN 406
Query: 144 ITTLTSL-ICSNLI 156
ITTLTS IC LI
Sbjct: 407 ITTLTSFPICICLI 448
Score = 70.1 bits (170), Expect(2) = 5e-13
Identities = 40/87 (45%), Positives = 51/87 (57%), Gaps = 1/87 (1%)
Frame = +1
Query: 441 LFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKSLGTNFDLPDLEVLNLT 499
+FPNL L LS I+ +GI +L C IRHL L C + FD+P LEVL+L+
Sbjct: 421 IFPNLHLLDLSDCSGISNQGIYHVLRRCCDIRHLYLADCSKVNLHRMIFDVPKLEVLDLS 600
Query: 500 NTEVDDEALYIISNRCPALLQLVLLRC 526
T VDD+ LY I+ C LLQL+L C
Sbjct: 601 YTRVDDKTLYAIAKSCRGLLQLLLQNC 681
Score = 22.3 bits (46), Expect(2) = 5e-13
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +2
Query: 529 ITDKGVMHVVNNC 541
+T+KGV VV NC
Sbjct: 689 VTEKGVERVVENC 727
>BE319841 weakly similar to GP|21592320|gb| unknown {Arabidopsis thaliana},
partial (10%)
Length = 363
Score = 124 bits (311), Expect = 1e-28
Identities = 68/121 (56%), Positives = 84/121 (69%)
Frame = +1
Query: 444 NLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEV 503
NLQ L LS I EEGI +L C IRHLNL+ S+ L +F++P LEVLNL+ T+V
Sbjct: 1 NLQLLDLSNCCRIFEEGIVQVLRMCCNIRHLNLSKCSIVRLEIDFEVPKLEVLNLSYTKV 180
Query: 504 DDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASM 563
DDEALY+IS C LL+L L C+ +T KGV HVV NCTQLR+I+L+GC V A VV+ M
Sbjct: 181 DDEALYMISKSCCGLLKLSLQDCNDVTKKGVKHVVENCTQLRKISLNGCFKVHANVVSLM 360
Query: 564 V 564
V
Sbjct: 361 V 363
>TC80264 similar to GP|21689699|gb|AAM67471.1 putative F-box family protein
AtFBL3 {Arabidopsis thaliana}, partial (4%)
Length = 1305
Score = 71.2 bits (173), Expect = 1e-12
Identities = 90/379 (23%), Positives = 162/379 (41%), Gaps = 7/379 (1%)
Frame = +3
Query: 222 LSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSITCTVTTGLEHLTSHFIDS 281
L+ + C L+++ L C +++ G+DL + L L ++ +T+ + S
Sbjct: 12 LAKIAVGCSKLEKLSLKWCLEISDLGIDLLSKKCFDLNFLDVSYL------KVTNESLRS 173
Query: 282 LLSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLV-LSYCPGYTYSGISFLLSKSKRIQ 340
+ SL L ++ G + D + PL + + +S C + SG+ ++S + ++
Sbjct: 174 IASLLKLEVFIMVGCYLVDDAGLQFLEKGCPLLKAIDVSRCNCVSPSGLLSVISGHEGLE 353
Query: 341 HLDLQYTDFLNDHCVAELSL----FLGDLLSLNLGNCRLLTVSTFFALI--TNCPSLTEI 394
++ HC++ELS L +L L++ + VS F I +NC SL E+
Sbjct: 354 QINA-------GHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVEL 512
Query: 395 NMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSY 454
+++ I T + +M + +L L + D I A PNL L L
Sbjct: 513 GLSKC-IGVTNM--GIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCD 683
Query: 455 NITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNR 514
+TE G+ + SC + L+LT S V+D AL +S R
Sbjct: 684 MVTEIGLYQIGSSCLMLEELDLTDCS---------------------GVNDIALKYLS-R 797
Query: 515 CPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIH 574
C L++L L C I+D G+ H+ NC +L E++L C + +A++ L ++
Sbjct: 798 CSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYRCVRIGDDGLAALTTGCNKLAMLN 977
Query: 575 VPPNFPLSDRNRKLFSRHG 593
+ ++D K S G
Sbjct: 978 LAYCNRITDAGLKCISNLG 1034
Score = 59.3 bits (142), Expect = 4e-09
Identities = 97/419 (23%), Positives = 171/419 (40%), Gaps = 4/419 (0%)
Frame = +3
Query: 71 KFCLTILYPALPVLPGLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLKLTSLNLSNQL 130
K+CL I + +L ++ L LD+SY ++L + S +LKL +
Sbjct: 60 KWCLEISDLGIDLLS---KKCFDLNFLDVSYLKVTNESLRSIAS---LLKLEVFIMVGCY 221
Query: 131 ILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHA 190
++ GL+ + L ++ S ++ + + + LE+++ + ++ A
Sbjct: 222 LVDDAGLQFLEKGCPLLKAIDVSRCNCVSPSGLLSVISGHEGLEQINAGH---CLSELSA 392
Query: 191 TFSTGLEALS-LALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVD 249
+ GL+ L L++I++ V +S +L + NCK L E+ L +C +T G+
Sbjct: 393 PLTNGLKNLKHLSVIRIDGVRVS------DFILQIIGSNCKSLVELGLSKCIGVTNMGI- 551
Query: 250 LALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAME 309
+ ++ LT+L +TC ++D +S+IA
Sbjct: 552 MQVVGCCNLTTLDLTCCRF-----------------------------VTDAAISTIANS 644
Query: 310 SLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLN 369
L L L C T G+ + S ++ LDL +ND + LS L+ L
Sbjct: 645 CPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR-CSKLVRLK 821
Query: 370 LGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAAC 429
LG C ++ + NCP LTE+++ R I + + L K L A C
Sbjct: 822 LGLCTNISDIGLAHIACNCPKLTELDLYRC----VRIGDDGLAALTTGCNKLAMLNLAYC 989
Query: 430 --LEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKSLG 485
+ D + + L L L NIT GI+ + SC+++ +LNL C L G
Sbjct: 990 NRITDAGLKCISNL-GELSDFELRGLSNITSIGIKAVAVSCKRLANLNLKHCEKLDDTG 1163
>TC86421 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max}, partial
(74%)
Length = 2856
Score = 68.6 bits (166), Expect = 7e-12
Identities = 97/415 (23%), Positives = 165/415 (39%), Gaps = 36/415 (8%)
Frame = +1
Query: 195 GLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQ 254
GL ++ KL K++L ++ L + K C L E+ L C + G+
Sbjct: 835 GLIEIANGCQKLEKLDLCKCPAISDKALITVAKKCPNLTELSLESCPSIRNEGLQAIGKF 1014
Query: 255 KPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIA------- 307
P L ++SI G + + F + L L T + L +SD L+ I
Sbjct: 1015 CPNLKAISIKDCAGVGDQGIAGLFSSTSLVL---TKVKLQALAVSDLSLAVIGHYGKTVT 1185
Query: 308 ---MESLP------------------LRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQY 346
+ LP L+ L ++ C G T GI + ++ + L
Sbjct: 1186 DLVLNFLPNVSERGFWVMGNANGLHKLKSLTIASCRGVTDVGIEAVGKGCPNLKSVHLHK 1365
Query: 347 TDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTI 406
FL+D+ + + L SL L C +T FF ++ NC + + + +
Sbjct: 1366 CAFLSDNGLISFTKAAISLESLQLEECHRITQFGFFGVLFNCGAKLKALSMISCFGIKDL 1545
Query: 407 PNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLE 466
L +SL + + + + + L P LQQ+ L+ +T+ G+ PLLE
Sbjct: 1546 DLELSPVSPCESLRSLSICNCPGFGNATLSVLGKLCPQLQQVELTGLKGVTDAGLLPLLE 1725
Query: 467 SCRK-IRHLNLT-CLSLK----SLGTNFDLPDLEVLNLTN-TEVDDEALYIISNRCPALL 519
S + +NL+ C++L S N LE+LNL + + +L I+ C L
Sbjct: 1726 SSEAGLVKVNLSGCVNLTDKVVSSLVNLHGWTLEILNLEGCINISNASLAAIAEHCQLLC 1905
Query: 520 QLVLLRCDYITDKGVMHVVN-NCTQLREINLDGCPNVQAKVVASMVVSRPSLRKI 573
L C I+D G+ + + L+ ++L GC V + S P+LRK+
Sbjct: 1906 DLDFSMCT-ISDSGITALAHAKQINLQILSLSGCTLVTDR-------SLPALRKL 2046
Score = 55.5 bits (132), Expect = 6e-08
Identities = 32/101 (31%), Positives = 58/101 (56%), Gaps = 1/101 (0%)
Frame = +1
Query: 476 LTCLSLKSLGTNFDLPDLEVLNLTN-TEVDDEALYIISNRCPALLQLVLLRCDYITDKGV 534
+T L LK++ + P L+ +L N + V DE L I+N C L +L L +C I+DK +
Sbjct: 745 VTTLGLKAVASG--CPSLKSFSLWNVSSVGDEGLIEIANGCQKLEKLDLCKCPAISDKAL 918
Query: 535 MHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHV 575
+ V C L E++L+ CP+++ + + ++ P+L+ I +
Sbjct: 919 ITVAKKCPNLTELSLESCPSIRNEGLQAIGKFCPNLKAISI 1041
Score = 44.3 bits (103), Expect = 1e-04
Identities = 50/261 (19%), Positives = 96/261 (36%), Gaps = 8/261 (3%)
Frame = +1
Query: 323 GYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFF 382
G T G+ + S ++ L + D + E++ L L+L C ++
Sbjct: 742 GVTTLGLKAVASGCPSLKSFSLWNVSSVGDEGLIEIANGCQKLEKLDLCKCPAISDKALI 921
Query: 383 ALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALF 442
+ CP+LTE++ L S + ++ +
Sbjct: 922 TVAKKCPNLTELS----------------------------LESCPSIRNEGLQAIGKFC 1017
Query: 443 PNLQQLHLSCSYNITEEGIRPLLESCR------KIRHLNLTCLSLKSLGTNFDLPDLEVL 496
PNL+ + + + ++GI L S K++ L ++ LSL +G VL
Sbjct: 1018PNLKAISIKDCAGVGDQGIAGLFSSTSLVLTKVKLQALAVSDLSLAVIGHYGKTVTDLVL 1197
Query: 497 NLTNTEVDDEALYII--SNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPN 554
N V + +++ +N L L + C +TD G+ V C L+ ++L C
Sbjct: 1198NFL-PNVSERGFWVMGNANGLHKLKSLTIASCRGVTDVGIEAVGKGCPNLKSVHLHKCAF 1374
Query: 555 VQAKVVASMVVSRPSLRKIHV 575
+ + S + SL + +
Sbjct: 1375LSDNGLISFTKAAISLESLQL 1437
>AW691925
Length = 649
Score = 66.6 bits (161), Expect = 3e-11
Identities = 42/119 (35%), Positives = 66/119 (55%), Gaps = 3/119 (2%)
Frame = +2
Query: 296 FRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTD-FLNDHC 354
FRIS LS + +C GYTY G+S LS+ ++I+HLDL +++D
Sbjct: 230 FRISSSHLSRYSR--------CTHFCLGYTYHGLSDFLSRRRQIRHLDLSGAPWYIHDEQ 385
Query: 355 VAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNI-QGT-TIPNSLM 411
+ L+ + D++S+NL +C LT + LIT C SL+E+ M T + +G+ T P L+
Sbjct: 386 IETLADLVTDIISINLTSCSKLTDYAVYILITTCRSLSELYMGDTKVGEGSETFPEQLL 562
>BF648657
Length = 675
Score = 63.5 bits (153), Expect = 2e-10
Identities = 34/69 (49%), Positives = 47/69 (67%)
Frame = +1
Query: 408 NSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLES 467
+SL+D + NPQ KSL+LA + L D+ +IMFA++F NLQ + LS NI++ I +L
Sbjct: 442 DSLLDFVFNPQLKSLYLAHNSWLRDEYLIMFASIFLNLQLIGLSYCDNISDNSICRVLNR 621
Query: 468 CRKIRHLNL 476
KIRHLNL
Sbjct: 622 WSKIRHLNL 648
Score = 35.8 bits (81), Expect(2) = 8e-07
Identities = 20/48 (41%), Positives = 27/48 (55%), Gaps = 1/48 (2%)
Frame = +3
Query: 226 FKNC-KFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSITCTVTTGLE 272
F C K + EVI+ +T G+ ALLQ+PTLTSLS + L+
Sbjct: 279 FSTCSKTVSEVIIFNSYHITNQGIASALLQRPTLTSLSFSTDFPNDLD 422
Score = 35.4 bits (80), Expect(2) = 8e-07
Identities = 27/76 (35%), Positives = 41/76 (53%), Gaps = 4/76 (5%)
Frame = +1
Query: 275 TSHFIDSLLSL---KGLTSLLLT-GFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGIS 330
TS FIDSLL L SL L + D++L A L L+ + LSYC + + I
Sbjct: 427 TSCFIDSLLDFVFNPQLKSLYLAHNSWLRDEYLIMFASIFLNLQLIGLSYCDNISDNSIC 606
Query: 331 FLLSKSKRIQHLDLQY 346
+L++ +I+HL+L +
Sbjct: 607 RVLNRWSKIRHLNLAH 654
>TC78335 similar to GP|16604366|gb|AAL24189.1 At1g21410/F24J8_17
{Arabidopsis thaliana}, partial (90%)
Length = 1520
Score = 62.8 bits (151), Expect = 4e-10
Identities = 61/210 (29%), Positives = 90/210 (42%), Gaps = 7/210 (3%)
Frame = +3
Query: 371 GNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACL 430
G CR S +F L SL+ N N N+ + +P + ++L L
Sbjct: 324 GVCRGWRDSIYFGLARL--SLSWCNKNMNNLVLSLVPKFA-------KLQTLILRQDKPQ 476
Query: 431 EDQNIIMFAALF-PNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFD 489
D N++ A F +LQ L LS S+ +T+ + + CR + LN++ S S
Sbjct: 477 LDDNVVGTIANFCHDLQILDLSKSFKLTDRSLYAIAHGCRDLTKLNISGCSAFSDNALAY 656
Query: 490 LPD----LEVLNLTNT--EVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQ 543
L L+VLNL D AL I + C L L L CD + D GVM + C
Sbjct: 657 LAGFCRKLKVLNLCGCVRAASDTALQAIGHYCNQLQSLNLGWCDKVGDVGVMSLAYGCPD 836
Query: 544 LREINLDGCPNVQAKVVASMVVSRPSLRKI 573
LR ++L GC + V ++ P LR +
Sbjct: 837 LRTVDLCGCVYITDDSVIALANGCPHLRSL 926
Score = 42.4 bits (98), Expect = 5e-04
Identities = 50/229 (21%), Positives = 91/229 (38%), Gaps = 2/229 (0%)
Frame = +3
Query: 292 LLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDL-QYTDFL 350
L F+++D+ L +IA L +L +S C ++ + +++L ++++ L+L
Sbjct: 537 LSKSFKLTDRSLYAIAHGCRDLTKLNISGCSAFSDNALAYLAGFCRKLKVLNLCGCVRAA 716
Query: 351 NDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSL 410
+D + + + L SLNLG C + +L CP L ++
Sbjct: 717 SDTALQAIGHYCNQLQSLNLGWCDKVGDVGVMSLAYGCPDLRTVD--------------- 851
Query: 411 MDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRK 470
L + D ++I A P+L+ L L NIT+ + L +S K
Sbjct: 852 -------------LCGCVYITDDSVIALANGCPHLRSLGLYFCKNITDNAMYSLAQSKVK 992
Query: 471 IRHLNLTCLSLKSLGTNFDLPDLEVLNLTN-TEVDDEALYIISNRCPAL 518
R S+ D L LN++ T + A+ + + PAL
Sbjct: 993 NRMWG-------SVKGGNDEDGLRTLNISQCTSLTPSAVQAVCDSSPAL 1118
>TC81462 similar to GP|21536497|gb|AAM60829.1 F-box protein family AtFBL4
{Arabidopsis thaliana}, partial (67%)
Length = 1452
Score = 59.7 bits (143), Expect = 3e-09
Identities = 40/147 (27%), Positives = 75/147 (50%), Gaps = 6/147 (4%)
Frame = +3
Query: 415 VNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHL 474
V KSL +A+ A + D ++ A+ +L+ L L + + +G+ + + C ++ L
Sbjct: 729 VGKSLKSLGVAACAKITDISMEAVASHCGSLETLSLDSEF-VHNQGVLAVAKGCPHLKSL 905
Query: 475 NLTCLSL-----KSLGTNFDLPDLEVLNLTNTE-VDDEALYIISNRCPALLQLVLLRCDY 528
L C++L K++G + LE+L L + + D+ L I N C L L L C +
Sbjct: 906 KLQCINLTDDALKAVGVS--CLSLELLALYSFQRFTDKGLRAIGNGCKKLKNLTLSDCYF 1079
Query: 529 ITDKGVMHVVNNCTQLREINLDGCPNV 555
++DKG+ + C +L + ++GC N+
Sbjct: 1080LSDKGLEAIATGCKELTHLEVNGCHNI 1160
Score = 43.9 bits (102), Expect = 2e-04
Identities = 33/105 (31%), Positives = 43/105 (40%)
Frame = +2
Query: 429 CLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNF 488
CL D +I A FP L++L L N+T G+ L C SLKS
Sbjct: 458 CLSDNGLIALADGFPKLEKLKLIWCSNVTSFGLSSLASK----------CASLKS----- 592
Query: 489 DLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKG 533
L+L V D+ L + RC L L L C+ +TD G
Sbjct: 593 -------LDLQGCYVGDQGLAAVGQRCKQLEDLNLRFCEGLTDTG 706
Score = 37.4 bits (85), Expect = 0.016
Identities = 53/244 (21%), Positives = 96/244 (38%), Gaps = 8/244 (3%)
Frame = +3
Query: 244 TIAGVDLALLQKPTLTSLSITCTVTTGLEHLT--SHFIDS---LLSLKG---LTSLLLTG 295
++ + +A K T S+ + LE L+ S F+ + L KG L SL L
Sbjct: 738 SLKSLGVAACAKITDISMEAVASHCGSLETLSLDSEFVHNQGVLAVAKGCPHLKSLKLQC 917
Query: 296 FRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCV 355
++D L ++ + L L L L +T G+ + + K++++L L FL+D +
Sbjct: 918 INLTDDALKAVGVSCLSLELLALYSFQRFTDKGLRAIGNGCKKLKNLTLSDCYFLSDKGL 1097
Query: 356 AELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLV 415
A+ T C LT + +N + GT +S+ +
Sbjct: 1098E--------------------------AIATGCKELTHLEVNGCHNIGTLGLDSVGKSCL 1199
Query: 416 NPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLN 475
+ L L + D ++ LQ LHL +I +E + + CR ++ L+
Sbjct: 1200H--LSELALLYCQRIGDLGLLQVGKGCQFLQALHLVDCSSIGDEAMCGIATGCRNLKKLH 1373
Query: 476 LTCL 479
+ L
Sbjct: 1374IPSL 1385
Score = 35.8 bits (81), Expect = 0.048
Identities = 36/148 (24%), Positives = 60/148 (40%)
Frame = +3
Query: 136 GLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTG 195
GLRA L +L S+ L+ + IA L L+ +N H + G
Sbjct: 1014 GLRAIGNGCKKLKNLTLSDCYFLSDKGLEAIATGCKELTHLE-------VNGCHNIGTLG 1172
Query: 196 LEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQK 255
L+++ + + L ++ L Y + L + K C+FLQ + L+ C + D A+
Sbjct: 1173 LDSVGKSCLHLSELALLYCQRIGDLGLLQVGKGCQFLQALHLVDCSSIG----DEAM--- 1331
Query: 256 PTLTSLSITCTVTTGLEHLTSHFIDSLL 283
C + TG +L I SL+
Sbjct: 1332 ---------CGIATGCRNLKKLHIPSLV 1388
Score = 34.7 bits (78), Expect = 0.11
Identities = 27/77 (35%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Frame = +2
Query: 478 CLSLKSLGTNFD-LPDLEVLNLT-NTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVM 535
CLS L D P LE L L + V L ++++C +L L L C Y+ D+G+
Sbjct: 458 CLSDNGLIALADGFPKLEKLKLIWCSNVTSFGLSSLASKCASLKSLDLQGC-YVGDQGLA 634
Query: 536 HVVNNCTQLREINLDGC 552
V C QL ++NL C
Sbjct: 635 AVGQRCKQLEDLNLRFC 685
>TC86288 similar to GP|2407790|gb|AAB70660.1| grr1 {Glycine max}, partial
(84%)
Length = 2900
Score = 59.7 bits (143), Expect = 3e-09
Identities = 89/386 (23%), Positives = 151/386 (39%), Gaps = 11/386 (2%)
Frame = +1
Query: 190 ATFS-TGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGV 248
AT S GL ++ ++ ++L ++ L + K+C L E+ + C + G+
Sbjct: 781 ATISDAGLIEIANGCHQIENLDLCKLPTISDKALIAVAKHCPNLTELSIESCPSIGNEGL 960
Query: 249 DLALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAM 308
P L S+SI + + + + LK LT L +SD L+ I
Sbjct: 961 HAIGKLCPNLRSVSIKNCPGVRDQGIAGLLCSASIILKKLT---LESLAVSDYSLAVIGQ 1131
Query: 309 ESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSL 368
+ LVL++ P T G + N H + +L+ SL
Sbjct: 1132 YGFVVTDLVLNFLPNVTEKG-----------------FWVMGNGHALQQLT-------SL 1239
Query: 369 NLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAA 428
+G CP +T+I ++ G PN K+ L +
Sbjct: 1240 TIG---------------LCPGVTDIGLHAV---GKGCPN----------VKNFQLRRCS 1335
Query: 429 CLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIR-PLLESCRKIRHLNL-TCLSLKSLGT 486
L D ++ F P++ L L + IT+ G+ +L K++ L L +C +K L
Sbjct: 1336 FLSDNGLVSFTKAAPSIVSLQLEECHRITQFGVAGAILNRGTKLKVLTLVSCYGIKDL-- 1509
Query: 487 NFDLP------DLEVLNLTNTE-VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVN 539
N +LP + L++ N V + L ++ CP L L L+ + IT+ G + ++
Sbjct: 1510 NLNLPAVPPCQKISSLSIRNCPGVGNFTLNVLGKLCPTLQCLELIGLEGITEPGFISLLQ 1689
Query: 540 NC-TQLREINLDGCPNVQAKVVASMV 564
L +NL GC N+ V SMV
Sbjct: 1690 RSKASLGNVNLSGCINLTDVGVLSMV 1767
Score = 48.9 bits (115), Expect = 5e-06
Identities = 32/151 (21%), Positives = 66/151 (43%)
Frame = +1
Query: 443 PNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTE 502
P+L+ L I++ G+ + C +I +L+L L
Sbjct: 748 PSLKSFTLWDVATISDAGLIEIANGCHQIENLDLCKLPT--------------------- 864
Query: 503 VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVAS 562
+ D+AL ++ CP L +L + C I ++G+ + C LR +++ CP V+ + +A
Sbjct: 865 ISDKALIAVAKHCPNLTELSIESCPSIGNEGLHAIGKLCPNLRSVSIKNCPGVRDQGIAG 1044
Query: 563 MVVSRPSLRKIHVPPNFPLSDRNRKLFSRHG 593
++ S + K + +SD + + ++G
Sbjct: 1045LLCSASIILKKLTLESLAVSDYSLAVIGQYG 1137
Score = 43.9 bits (102), Expect = 2e-04
Identities = 49/191 (25%), Positives = 82/191 (42%), Gaps = 9/191 (4%)
Frame = +1
Query: 374 RLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNP--QFKSLFLASAACLE 431
R LT A+ CPSL + TI ++ + + N Q ++L L +
Sbjct: 703 RALTDVGLKAVAHGCPSLKSFTLWDV----ATISDAGLIEIANGCHQIENLDLCKLPTIS 870
Query: 432 DQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNL-TCLSLKSLGTNFDL 490
D+ +I A PNL +L + +I EG+ + + C +R +++ C ++ G L
Sbjct: 871 DKALIAVAKHCPNLTELSIESCPSIGNEGLHAIGKLCPNLRSVSIKNCPGVRDQGIAGLL 1050
Query: 491 PD----LEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKG--VMHVVNNCTQL 544
L+ L L + V D +L +I + LVL +T+KG VM + QL
Sbjct: 1051CSASIILKKLTLESLAVSDYSLAVIGQYGFVVTDLVLNFLPNVTEKGFWVMGNGHALQQL 1230
Query: 545 REINLDGCPNV 555
+ + CP V
Sbjct: 1231TSLTIGLCPGV 1263
Score = 43.5 bits (101), Expect = 2e-04
Identities = 46/214 (21%), Positives = 92/214 (42%), Gaps = 9/214 (4%)
Frame = +1
Query: 369 NLGNCRLLTVS--TFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLAS 426
NL C+L T+S A+ +CP+LTE+++ G +++ + P +S+ + +
Sbjct: 838 NLDLCKLPTISDKALIAVAKHCPNLTELSIESCPSIGNEGLHAIGK--LCPNLRSVSIKN 1011
Query: 427 AACLEDQNII-MFAALFPNLQQLHLSC----SYNITEEGIRPLLESCRKIRHL-NLTCLS 480
+ DQ I + + L++L L Y++ G + + + L N+T
Sbjct: 1012 CPGVRDQGIAGLLCSASIILKKLTLESLAVSDYSLAVIGQYGFVVTDLVLNFLPNVTEKG 1191
Query: 481 LKSLGTNFDLPDLEVLNLTNTE-VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVN 539
+G L L L + V D L+ + CP + L RC +++D G++
Sbjct: 1192 FWVMGNGHALQQLTSLTIGLCPGVTDIGLHAVGKGCPNVKNFQLRRCSFLSDNGLVSFTK 1371
Query: 540 NCTQLREINLDGCPNVQAKVVASMVVSRPSLRKI 573
+ + L+ C + VA +++R + K+
Sbjct: 1372 AAPSIVSLQLEECHRITQFGVAGAILNRGTKLKV 1473
Score = 40.4 bits (93), Expect = 0.002
Identities = 60/275 (21%), Positives = 119/275 (42%), Gaps = 3/275 (1%)
Frame = +1
Query: 195 GLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQ 254
GL A+ ++ L +L+ L K + + L C ++T GV A+L
Sbjct: 1273 GLHAVGKGCPNVKNFQLRRCSFLSDNGLVSFTKAAPSIVSLQLEECHRITQFGVAGAILN 1452
Query: 255 KPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMESLP-L 313
+ T + +T G++ L + + ++ + ++SL + F ++ + P L
Sbjct: 1453 RGTKLKV-LTLVSCYGIKDLNLN-LPAVPPCQKISSLSIRNCPGVGNFTLNVLGKLCPTL 1626
Query: 314 RRLVLSYCPGYTYSGISFLLSKSKR-IQHLDLQYTDFLNDHCVAEL-SLFLGDLLSLNLG 371
+ L L G T G LL +SK + +++L L D V + L L LNL
Sbjct: 1627 QCLELIGLEGITEPGFISLLQRSKASLGNVNLSGCINLTDVGVLSMVKLHCSTLGVLNLN 1806
Query: 372 NCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLE 431
C+ + ++ A+ NC L++++++ I I S + R V L LA + +
Sbjct: 1807 GCKKVGDASLTAIADNCIVLSDLDVSECAITDAGI--SALTRGVLFNLDVLSLAGCSLVS 1980
Query: 432 DQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLE 466
++++ L +L+ L++ +I+ + L+E
Sbjct: 1981 NKSLSALKKLGDSLEGLNIKNCKSISSRTVNKLVE 2085
Score = 38.9 bits (89), Expect = 0.006
Identities = 17/71 (23%), Positives = 36/71 (49%)
Frame = +1
Query: 505 DEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMV 564
D L +++ CP+L L I+D G++ + N C Q+ ++L P + K + ++
Sbjct: 715 DVGLKAVAHGCPSLKSFTLWDVATISDAGLIEIANGCHQIENLDLCKLPTISDKALIAVA 894
Query: 565 VSRPSLRKIHV 575
P+L ++ +
Sbjct: 895 KHCPNLTELSI 927
>TC79219 similar to GP|15810000|gb|AAL06927.1 AT5g23340/MKD15_20
{Arabidopsis thaliana}, partial (78%)
Length = 1619
Score = 59.7 bits (143), Expect = 3e-09
Identities = 64/255 (25%), Positives = 111/255 (43%), Gaps = 38/255 (14%)
Frame = +2
Query: 339 IQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNR 398
+ LD+ Y L D ++ ++ DL L+L CR +T S AL NC +L E+
Sbjct: 491 LHSLDVSYCRKLTDKGLSAVAKGCCDLRILHLTGCRFVTDSILEALSKNCRNLEEL---- 658
Query: 399 TNIQGTT--IPNSLMD----------------------------RLVNPQFKSLFLASAA 428
+QG T N LM + K+L L
Sbjct: 659 -VLQGCTSITDNGLMSLASGCQRIKFLDINKCSTVSDVGVSSICNACSSSLKTLKLLDCY 835
Query: 429 CLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCR-KIRHLNLT-CLSLKSLGT 486
+ D++I+ A NL+ L + +++ + I+ L +CR K+++L + CL++
Sbjct: 836 RIGDKSILSLAKFCDNLETLIIGGCRDVSNDAIKLLATACRNKLKNLRMDWCLNVSDSSL 1015
Query: 487 NFDLP---DLEVLNL-TNTEVDDEALYIISNRCPALLQLVL--LRCDYITDKGVMHVVNN 540
+ L +LE L++ EV D A + ISN P L +L C IT G+ ++
Sbjct: 1016SCILSQCRNLEALDIGCCEEVTDTAFHHISNEEPGLSLKILKVSNCPKITVVGIGILLGK 1195
Query: 541 CTQLREINLDGCPNV 555
C+ L+ +++ CP++
Sbjct: 1196CSYLKYLDVRSCPHI 1240
Score = 46.6 bits (109), Expect = 3e-05
Identities = 32/119 (26%), Positives = 55/119 (45%)
Frame = +2
Query: 445 LQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVD 504
L L +S +T++G+ + + C +R L+LT G F V
Sbjct: 491 LHSLDVSYCRKLTDKGLSAVAKGCCDLRILHLT-------GCRF--------------VT 607
Query: 505 DEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASM 563
D L +S C L +LVL C ITD G+M + + C +++ ++++ C V V+S+
Sbjct: 608 DSILEALSKNCRNLEELVLQGCTSITDNGLMSLASGCQRIKFLDINKCSTVSDVGVSSI 784
Score = 34.7 bits (78), Expect = 0.11
Identities = 19/66 (28%), Positives = 32/66 (47%)
Frame = +2
Query: 498 LTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQA 557
+T E+ D + I + L L + C +TDKG+ V C LR ++L GC V
Sbjct: 431 ITVKELPDVGMKAIGDGLSLLHSLDVSYCRKLTDKGLSAVAKGCCDLRILHLTGCRFVTD 610
Query: 558 KVVASM 563
++ ++
Sbjct: 611 SILEAL 628
Score = 34.3 bits (77), Expect = 0.14
Identities = 21/78 (26%), Positives = 40/78 (50%), Gaps = 1/78 (1%)
Frame = +2
Query: 479 LSLKSLGTNFDLPDLEVLNLTNT-EVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHV 537
+ +K++G L L L+++ ++ D+ L ++ C L L L C ++TD + +
Sbjct: 455 VGMKAIGDGLSL--LHSLDVSYCRKLTDKGLSAVAKGCCDLRILHLTGCRFVTDSILEAL 628
Query: 538 VNNCTQLREINLDGCPNV 555
NC L E+ L GC ++
Sbjct: 629 SKNCRNLEELVLQGCTSI 682
>TC84223 weakly similar to GP|18252175|gb|AAL61920.1 unknown protein
{Arabidopsis thaliana}, partial (90%)
Length = 790
Score = 56.6 bits (135), Expect = 3e-08
Identities = 34/136 (25%), Positives = 66/136 (48%)
Frame = +3
Query: 436 IMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEV 495
I +A NL + L IT+EG+ L+ + +RH
Sbjct: 396 ISYAKCINNLTSISLWGLTGITDEGVVKLISRTKSLRH---------------------- 509
Query: 496 LNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNV 555
LN+ T + DE+L+ I+ CP + +VL C ++T+ G++ +V+ C +L+ +N+ G V
Sbjct: 510 LNVGGTFITDESLFAIARSCPKMETIVLWSCRHVTENGLIALVDQCLKLKSMNVWGL-RV 686
Query: 556 QAKVVASMVVSRPSLR 571
+ ++++ P+L+
Sbjct: 687 PVDCLNTLLIMSPTLQ 734
>BQ122644
Length = 444
Score = 55.8 bits (133), Expect = 4e-08
Identities = 45/108 (41%), Positives = 60/108 (54%), Gaps = 4/108 (3%)
Frame = +3
Query: 446 QQLHLSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKSLGTNFDLPDLEVLNLTNTEVD 504
Q L LS +++EEGI +L C IRHLNLT C LK NF++P LEVL+L+ T V+
Sbjct: 129 QLLDLSHCKDVSEEGIVHVLMICCNIRHLNLTRCSRLKLHTLNFEVPKLEVLDLSFTRVE 308
Query: 505 DEALYIISN--RCPALLQLVLLRCD-YITDKGVMHVVNNCTQLREINL 549
L + R A + +L RC ++D CTQLRE+NL
Sbjct: 309 *ITLLDLKEL*RAFATVT*IL*RCHRQMSDA----CG*KCTQLREVNL 440
>TC86245 weakly similar to GP|9279671|dbj|BAB01228.1 transport inhibitor
response-like protein {Arabidopsis thaliana}, partial
(80%)
Length = 2260
Score = 53.5 bits (127), Expect = 2e-07
Identities = 75/300 (25%), Positives = 127/300 (42%), Gaps = 19/300 (6%)
Frame = +3
Query: 288 LTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYT 347
L L L +SD+ L +A + L L C G++ G++ + + K + LD+Q
Sbjct: 810 LEELRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCKNLTELDIQ-E 986
Query: 348 DFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFF----ALITNCPSLTEINMNRTNIQG 403
+ ++D LS F SL + N L+ F L+ C SL + +N+
Sbjct: 987 NGVDDKSGNWLSCFPESFTSLEILNFANLSNDVNFDALEKLVARCNSLKTLKVNK----- 1151
Query: 404 TTIPNSLMDRLV--NPQFKSLFLAS-AACLEDQNIIMFAALFPNLQQLH-LSCSYNITEE 459
++ + RL+ PQ L S + L Q F N + LH LS + + +
Sbjct: 1152 -SVTLEQLQRLLVRAPQLCELGTGSFSQELTGQQYSELERAFNNCRSLHTLSGLWVASAQ 1328
Query: 460 GIRPLLESCRKIRHLNLTCLSLKSLGTNFDL---PDLEVLNLTNTEVDDEALYIISNRCP 516
+ L C + LN + L S G + L P+L L + +T V+D+ L + + CP
Sbjct: 1329 YHQVLYPVCTNLTFLNFSYAPLDSEGLSKLLVRCPNLRRLWVLDT-VEDKGLEAVGSYCP 1505
Query: 517 ALLQLVLLRCD--------YITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRP 568
L +L + D +T+ G + V C +L + L C + VA++V + P
Sbjct: 1506 LLEELRVFPGDPFEEGAAHGVTESGFIAVSEGCRKLHYV-LYFCRQMTNAAVATVVENCP 1682
Score = 38.5 bits (88), Expect = 0.007
Identities = 21/59 (35%), Positives = 29/59 (48%)
Frame = +3
Query: 491 PDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINL 549
P LE L L V DE+L ++ P L LL CD + G+ V NC L E+++
Sbjct: 804 PFLEELRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCKNLTELDI 980
>TC86244 weakly similar to PIR|T48087|T48087 transport inhibitor response
protein TIR1 [imported] - Arabidopsis thaliana, partial
(48%)
Length = 2369
Score = 51.6 bits (122), Expect = 8e-07
Identities = 62/264 (23%), Positives = 109/264 (40%), Gaps = 13/264 (4%)
Frame = +1
Query: 288 LTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYT 347
L L L ++D+ L +A + L L C G++ G++ + + K + LD+Q
Sbjct: 538 LEELRLKRMVVTDESLEFLAFSFHNFKALSLLSCEGFSTDGLAAVAANCKNLTELDIQEN 717
Query: 348 DFLNDHCVAELSLFLGDLLSLNLGNCRLL----TVSTFFALITNCPSLTEINMNRTNIQG 403
D ++D LS F SL + N L + L+ C SL + +N++
Sbjct: 718 D-IDDKSGDWLSCFPESFTSLEVLNFANLNNDVNIDALEKLVGRCKSLKTLKVNKS---- 882
Query: 404 TTIPNSLMDRLVNPQFKSLFLAS-AACLEDQNIIMFAALFPNLQQLHLSCSYNITEEG-- 460
T+ ++ PQ L S + L Q + + F N + LH +
Sbjct: 883 VTLEQFQRLLVLAPQLCELGSGSFSQDLTCQQYLELESAFKNCKSLHTLSGLWVASASAQ 1062
Query: 461 ---IRPLLESCRKIRHLNLTCLSLKSLGTN---FDLPDLEVLNLTNTEVDDEALYIISNR 514
++ L +C + LN + + S P+L L + +T V+D+ L + +
Sbjct: 1063YIQLQVLYSACTNLTFLNFSYALVDSEDLTDLLVHCPNLRRLWVVDT-VEDKGLEAVGSY 1239
Query: 515 CPALLQLVLLRCDYITDKGVMHVV 538
CP L +L + D D+GV+H V
Sbjct: 1240CPLLEELRVFPADPF-DEGVVHGV 1308
>TC85267 similar to GP|9279671|dbj|BAB01228.1 transport inhibitor
response-like protein {Arabidopsis thaliana}, partial
(48%)
Length = 1795
Score = 48.1 bits (113), Expect = 9e-06
Identities = 48/184 (26%), Positives = 83/184 (45%), Gaps = 7/184 (3%)
Frame = +1
Query: 250 LALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLK-GLTSLLLTGFRISDQFLSSIAM 308
L L KP S+ V G +I++L K GL L L +SD+ L ++
Sbjct: 1111 LTLKGKPHFADFSL---VPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSR 1281
Query: 309 ESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLF---LGDL 365
+ + LVL C G+T G++ + + + ++ LDLQ + + DH LS F L
Sbjct: 1282 SFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENE-VEDHKGQWLSCFPESCTSL 1458
Query: 366 LSLNLGNCR-LLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVN--PQFKSL 422
+SLN + + + L++ P+L + +NR ++P + R++ PQ L
Sbjct: 1459 VSLNFACLKGDINLGALERLVSRSPNLKSLRLNR------SVPVDALQRILTRAPQLMDL 1620
Query: 423 FLAS 426
+ S
Sbjct: 1621 GIGS 1632
Score = 40.0 bits (92), Expect = 0.003
Identities = 22/57 (38%), Positives = 30/57 (52%)
Frame = +1
Query: 493 LEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINL 549
LE L L V DE+L ++S LVL+ C+ T G+ V NC LRE++L
Sbjct: 1219 LEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDL 1389
>TC76362 similar to GP|10716949|gb|AAG21977.1 SKP1 interacting partner 2
{Arabidopsis thaliana}, partial (57%)
Length = 1307
Score = 46.2 bits (108), Expect = 4e-05
Identities = 24/81 (29%), Positives = 47/81 (57%), Gaps = 4/81 (4%)
Frame = +1
Query: 497 NLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGC---- 552
N +T ++D+AL +IS RC L +L L C +T+ G++ + NC L+++++ C
Sbjct: 451 NKKSTSINDDALILISLRCKNLTRLKLRACREVTEIGMLGLARNCKNLKKLSVVSCLFGV 630
Query: 553 PNVQAKVVASMVVSRPSLRKI 573
++A V S V+ S++++
Sbjct: 631 KGIRAVVDNSNVLEELSVKRL 693
Score = 31.6 bits (70), Expect = 0.91
Identities = 26/111 (23%), Positives = 44/111 (39%)
Frame = +1
Query: 445 LQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVD 504
L+ LHL + ++ G+ + E CR ++ L++ +G
Sbjct: 988 LETLHLVKTPECSDHGLVVIAERCRMLKKLHIDGWRTNRIG------------------- 1110
Query: 505 DEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNV 555
DE L ++ CP L +LVL+ Y T + + + C L L G V
Sbjct: 1111 DEGLVSVARNCPNLQELVLIAM-YPTSLSLGAIASCCLGLERFALCGIGTV 1260
>TC85594 similar to PIR|D96512|D96512 hypothetical protein F2G19.16
[imported] - Arabidopsis thaliana, partial (38%)
Length = 1334
Score = 46.2 bits (108), Expect = 4e-05
Identities = 33/138 (23%), Positives = 64/138 (45%), Gaps = 6/138 (4%)
Frame = +1
Query: 444 NLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDL------PDLEVLN 497
NL+ LHL + T+ G+ + E C+ +R L++ +G + P+L+ L
Sbjct: 136 NLEILHLVKTPECTDMGLVAIAERCKLLRKLHIDGWKANRIGDEGLIAVAKFCPNLQELV 315
Query: 498 LTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQA 557
L +L ++++ CP L +L L D + D + + C L+++ + CP V
Sbjct: 316 LIGVNPTRVSLEMLASNCPNLERLALCASDTVGDPEISCIAAKCLALKKLCIKSCP-VSD 492
Query: 558 KVVASMVVSRPSLRKIHV 575
+ ++ P+L K+ V
Sbjct: 493 LGMEALANGCPNLVKVKV 546
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,718,504
Number of Sequences: 36976
Number of extensions: 325686
Number of successful extensions: 2006
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 1882
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1951
length of query: 598
length of database: 9,014,727
effective HSP length: 102
effective length of query: 496
effective length of database: 5,243,175
effective search space: 2600614800
effective search space used: 2600614800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140915.5


