

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.6 - phase: 0
(831 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
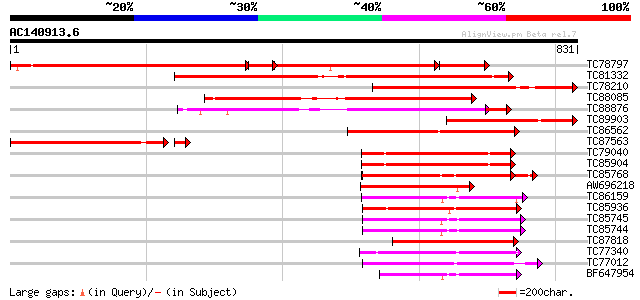
Score E
Sequences producing significant alignments: (bits) Value
TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 486 0.0
TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 570 e-163
TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Ar... 377 e-104
TC88085 similar to GP|20259341|gb|AAM13995.1 unknown protein {Ar... 335 4e-92
TC88876 similar to PIR|G96537|G96537 hypothetical protein F2J10.... 290 1e-84
TC89903 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22... 271 8e-73
TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regu... 259 3e-69
TC87563 similar to GP|13027363|dbj|BAB32724. hypothetical protei... 239 5e-66
TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase ... 187 1e-47
TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [i... 187 1e-47
TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle pr... 182 4e-46
AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33... 167 1e-41
TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AA... 159 4e-39
TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidops... 156 4e-38
TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome re... 154 1e-37
TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15... 154 1e-37
TC87818 similar to GP|20197082|gb|AAC26698.2 putative katanin {A... 152 7e-37
TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog ... 147 2e-35
TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulator... 144 1e-34
BF647954 homologue to GP|11094192|db 26S proteasome regulatory p... 143 3e-34
>TC78797 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22
{Arabidopsis thaliana}, partial (74%)
Length = 2270
Score = 486 bits (1252), Expect(4) = 0.0
Identities = 261/362 (72%), Positives = 300/362 (82%), Gaps = 10/362 (2%)
Frame = +2
Query: 1 MEQKGMLISA---ALSVGVGVGLGLA-SGQTMFK--PNTYSSSSNALTPDKIENEMLRLV 54
ME K +LISA + VGVGVG+GLA SGQ + K PN YS SSNA+T DKIE+EMLR +
Sbjct: 104 MEHKSILISALSVGVGVGVGVGIGLANSGQNVSKWGPN-YSFSSNAVTADKIEHEMLRQI 280
Query: 55 VDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAEL 114
VDGRESNVTFD FPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAEL
Sbjct: 281 VDGRESNVTFDKFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAEL 460
Query: 115 YQQVLAKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSETALARLSDLFGS 174
YQQVLAKAL HYFEAKLLL D+TDFSLKIQ +YG+ N E SF RSTSE+ L +LSD+FGS
Sbjct: 461 YQQVLAKALAHYFEAKLLLLDLTDFSLKIQGKYGTGNKEFSFKRSTSESTLEKLSDIFGS 640
Query: 175 FALFPQREENQ--GKIHRQSSGSDLRQMEAEGSYS--KLRRNASASANISSIGLQSNPTN 230
F++F QREE + GK++RQSSG DL+ M E S + KLRRNAS+++NIS + QSNPTN
Sbjct: 641 FSIFSQREEPKVSGKMNRQSSGMDLQSMGDEASCNPPKLRRNASSTSNISGLTSQSNPTN 820
Query: 231 SAPGKHITGWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLT 290
AP K W FDEK+LIQ+LYKVLL VSKTYPIVLY+RD D+LL RSQ+IY +FQ ML
Sbjct: 821 PAPLKRTASWSFDEKLLIQSLYKVLLSVSKTYPIVLYLRDVDRLLSRSQKIYNMFQKMLK 1000
Query: 291 KLSGPILIIGSRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKI 350
+LSGPILI+GSR+LDSGNE + +DE LT LFPY+I+I+PPEDES LVSWKSQ E DMK I
Sbjct: 1001RLSGPILILGSRVLDSGNEFEELDEKLTVLFPYDIKIRPPEDESHLVSWKSQLEEDMKMI 1180
Query: 351 QI 352
Q+
Sbjct: 1181QV 1186
Score = 357 bits (915), Expect(4) = 0.0
Identities = 194/251 (77%), Positives = 207/251 (82%), Gaps = 9/251 (3%)
Frame = +1
Query: 389 EEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSE 448
+ ++VSAIS+H+MKNK+ EYRNGKLII CNSLSHALGIF+ GKF RD+ KLE QA+ SE
Sbjct: 1297 KRLLVSAISHHLMKNKDLEYRNGKLIISCNSLSHALGIFRKGKFSGRDTSKLEDQALKSE 1476
Query: 449 KKEEGAAV-KPEGKTESPAPA------VKTEAEIPTSVRKTDGENSVPASKAEV--PDNE 499
V KP K ES APA K EAEI TSV K E V ASKA PDNE
Sbjct: 1477 IPSVVETVAKPAVKAESAAPADGAAPVKKAEAEISTSVAKAGAEKPVAASKASEVPPDNE 1656
Query: 500 FEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLF 559
FEKRIRPEVIPANEI VTFSDIGAL+ETK+SLQELVMLPLRRPDLF GGLLKPCRGILLF
Sbjct: 1657 FEKRIRPEVIPANEIDVTFSDIGALEETKESLQELVMLPLRRPDLFTGGLLKPCRGILLF 1836
Query: 560 GPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVD 619
GPPGTGKTMLAKAIA EAGASFINVSMSTITSKWFGEDEKNVRALFTLA+KVSPTIIFVD
Sbjct: 1837 GPPGTGKTMLAKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVD 2016
Query: 620 EVDSMLGQRTR 630
EVDSMLGQRT+
Sbjct: 2017 EVDSMLGQRTK 2049
Score = 122 bits (305), Expect(4) = 0.0
Identities = 60/73 (82%), Positives = 67/73 (91%)
Frame = +3
Query: 631 VGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSA 690
+G +AMRKIKNEFMS+WDGLT+K +RILVLAATN+PFDLDEAIIRRFERRIMVGLPS
Sbjct: 2052 LGSMKAMRKIKNEFMSHWDGLTTKQGERILVLAATNKPFDLDEAIIRRFERRIMVGLPSV 2231
Query: 691 ENRENILRTLLAK 703
ENRE ILRTLL+K
Sbjct: 2232 ENREKILRTLLSK 2270
Score = 77.8 bits (190), Expect(4) = 0.0
Identities = 37/42 (88%), Positives = 38/42 (90%)
Frame = +3
Query: 351 QIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEII 392
+ QDNKNHI EVLAANDLDC DLDSICV DTMVLSNYIEEII
Sbjct: 1182 KFQDNKNHITEVLAANDLDCDDLDSICVEDTMVLSNYIEEII 1307
Score = 31.6 bits (70), Expect = 1.3
Identities = 26/94 (27%), Positives = 43/94 (45%)
Frame = +1
Query: 30 KPNTYSSSSNALTPDKIENEMLRLVVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKH 89
KP S +S ++ E + V+ E +VTF + E+T+ L + L+
Sbjct: 1609 KPVAASKASEVPPDNEFEKRIRPEVIPANEIDVTFSDIGAL--EETKESLQELVMLPLRR 1782
Query: 90 AEVSKYTRNLAPASRTILLSGPAELYQQVLAKAL 123
++ +T L R ILL GP + +LAKA+
Sbjct: 1783 PDL--FTGGLLKPCRGILLFGPPGTGKTMLAKAI 1878
>TC81332 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22
{Arabidopsis thaliana}, partial (39%)
Length = 1514
Score = 570 bits (1469), Expect = e-163
Identities = 297/498 (59%), Positives = 370/498 (73%), Gaps = 1/498 (0%)
Frame = +1
Query: 242 FDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILIIGS 301
FDEK+ + +LYKVL+ +S ++LY+++ +K+ S R+Y+LFQ L KLSG +LI+GS
Sbjct: 79 FDEKLFLHSLYKVLVSISXAGSVILYIKNVEKVFLGSPRMYRLFQKTLNKLSGSVLILGS 258
Query: 302 RILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIME 361
R D C +V+E LT LFPYNIEI PP+DE+ L WKSQ + MKK ++D HI E
Sbjct: 259 RPYDLKYNCTKVNEKLTMLFPYNIEITPPQDETHLKIWKSQLKKAMKKTHLKDYTTHIAE 438
Query: 362 VLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLS 421
VLAANDL C DLD++ D +LSN EE++ SAI +H+ K P+YRNG LII SL
Sbjct: 439 VLAANDLYCDDLDTVDHNDMTILSNQTEEVVASAIFHHLKDAKNPKYRNGILIISAKSLR 618
Query: 422 HALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRK 481
H L +FQ G+ ++D+ K + ++ + ++E KP+ K
Sbjct: 619 HVLSLFQEGESSEKDNKKTKKESKRDDSRKE----KPKES-------------------K 729
Query: 482 TDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRR 541
DG+ A K++ PDN FE+ IR E+IPANEI VTFSDIGALD+ K+SLQE VMLPLRR
Sbjct: 730 KDGDIKASA-KSDSPDNAFEECIRQELIPANEIKVTFSDIGALDDVKESLQEAVMLPLRR 906
Query: 542 PDLFEG-GLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKN 600
PDLF+G G+LKPC+G+LLFGPPGTGKTMLAKAIANEAGASFINVS STITS W G+ EKN
Sbjct: 907 PDLFKGDGVLKPCKGVLLFGPPGTGKTMLAKAIANEAGASFINVSPSTITSMWQGQSEKN 1086
Query: 601 VRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRIL 660
VRALF+LAAKV+PTIIF+DEVDSMLGQR+ EH +MR++KNEFMS WDGL SK +++I
Sbjct: 1087VRALFSLAAKVAPTIIFIDEVDSMLGQRSSTREHSSMRRVKNEFMSRWDGLLSKPDEKIT 1266
Query: 661 VLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTE 720
VLAATN PFDLDEAIIRRF+RRIMVGLPSA+NRE IL+T+LAKEK ++F+EL+TMTE
Sbjct: 1267VLAATNMPFDLDEAIIRRFQRRIMVGLPSADNRETILKTILAKEKSE-NMNFEELSTMTE 1443
Query: 721 GYSGSDLKNLCTTAAYRP 738
GYSGSDLKNLC TAAYRP
Sbjct: 1444GYSGSDLKNLCMTAAYRP 1497
>TC78210 similar to GP|6056413|gb|AAF02877.1| Unknown protein {Arabidopsis
thaliana}, partial (23%)
Length = 1254
Score = 377 bits (968), Expect = e-104
Identities = 191/301 (63%), Positives = 236/301 (77%), Gaps = 1/301 (0%)
Frame = +3
Query: 532 QELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTIT 590
+ELVMLPL+RP+LF +G L KPC+GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+IT
Sbjct: 3 KELVMLPLKRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSIT 182
Query: 591 SKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDG 650
SKWFGE EK V+A+F+LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDG
Sbjct: 183 SKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDG 362
Query: 651 LTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGL 710
L +K ++RILVLAATNRPFDLDEA+IRR RR+MV LP A NR ILR +LAKE + +
Sbjct: 363 LRTKEKERILVLAATNRPFDLDEAVIRRLPRRLMVDLPDAPNRGKILRVILAKEDLAADV 542
Query: 711 DFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKE 770
D + +A MT+GYSGSDLKNLC TAA+ P+RE+ ++K+ K K A +N K
Sbjct: 543 DLEAIANMTDGYSGSDLKNLCVTAAHCPIREI----LEKEKKDKSLALAEN-------KP 689
Query: 771 EVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYF 830
E E +RPL M+DF+ A QV AS ++E MNEL+QWNDLYGEGGSRK + LSYF
Sbjct: 690 EPELCSSADIRPLKMEDFRYAHEQVCASVSSESTNMNELQQWNDLYGEGGSRKMKSLSYF 869
Query: 831 L 831
+
Sbjct: 870 M 872
>TC88085 similar to GP|20259341|gb|AAM13995.1 unknown protein {Arabidopsis
thaliana}, partial (29%)
Length = 1261
Score = 335 bits (859), Expect = 4e-92
Identities = 187/399 (46%), Positives = 250/399 (61%), Gaps = 1/399 (0%)
Frame = +1
Query: 286 QTMLTKLSGPILIIGSRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEA 345
QT L L+ P +R+ D E +V + L FP + I+ P+DE+ L WK E
Sbjct: 220 QTALLDLAFPDNF--TRLHDRSKETPKVMKQLNRFFPNKVTIQLPQDEALLSDWKQHLER 393
Query: 346 DMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKE 405
D++ ++ Q N I VL LDC +L+++ + D + + +E+II AISYH M + E
Sbjct: 394 DVETMKAQSNVVSIRLVLNKFGLDCPELETLSIKDQTLTTENVEKIIGWAISYHFMHSSE 573
Query: 406 PEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESP 465
K +I S+ + I Q + +E K
Sbjct: 574 ASTEESKPVISAESIQYGFNIL---------------QGIQNENK--------------- 663
Query: 466 APAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALD 525
SV+K+ K V +NEFEK++ +VIP +IGV+F+DIGAL+
Sbjct: 664 ------------SVKKS--------LKDVVTENEFEKKLLGDVIPPTDIGVSFNDIGALE 783
Query: 526 ETKDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINV 584
KD+L+ELVMLPL+RP+LF +G L KPC+GILLFGPPGTGKTMLAKA+A EAGA+FIN+
Sbjct: 784 NVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATEAGANFINI 963
Query: 585 SMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEF 644
SMS+ITSKWFGE EK V+A+F+LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEF
Sbjct: 964 SMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEF 1143
Query: 645 MSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRI 683
M NWDGL +K +R+LVLAATNRPFDLDEA+IRR RR+
Sbjct: 1144MVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRL 1260
>TC88876 similar to PIR|G96537|G96537 hypothetical protein F2J10.1
[imported] - Arabidopsis thaliana, partial (72%)
Length = 1572
Score = 290 bits (742), Expect(2) = 1e-84
Identities = 181/499 (36%), Positives = 260/499 (51%), Gaps = 40/499 (8%)
Frame = +2
Query: 246 ILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRS------QRIYKLFQTMLTKLSGPILII 299
I ++ L +VL + P+++Y D+ + L +S + + M +L GP+++I
Sbjct: 146 IAVEALCEVL---NSKRPLIVYFPDSSQWLHKSVPKSNRNEFFHKVEEMFDRLYGPVVLI 316
Query: 300 -GSRILDSGNECKRVDEML--------------------------------TSLFPYNIE 326
G + SG++ K M+ LF +
Sbjct: 317 CGQNKVHSGSKEKEKFTMILPNFGRVAKLPLSLKHLTDGFKGGKTSEEDDINKLFSNVLS 496
Query: 327 IKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLSN 386
+ PP++E+ +K Q E D K + + N N + +VL + L C DL + ++
Sbjct: 497 VHPPKEENLQTVFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCTDLLHVNTDGIVITKQ 676
Query: 387 YIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVT 446
E+++ A ++++ P + +L IP SL A+ +++
Sbjct: 677 KAEKLVGWAKNHYLSSCLLPSIKGERLCIPRESLEIAIS-------------RMKGMETM 817
Query: 447 SEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRP 506
S K + + +EFE
Sbjct: 818 SRKSSQNLK--------------------------------------NLAKDEFESNFVS 883
Query: 507 EVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFE-GGLLKPCRGILLFGPPGTG 565
V+ EIGV F DIGAL++ K +LQELV+LP+RRP+LF G LL+PC+GILLFGPPGTG
Sbjct: 884 AVVAPGEIGVKFDDIGALEDVKKALQELVILPMRRPELFSHGNLLRPCKGILLFGPPGTG 1063
Query: 566 KTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSML 625
KT+LAKA+A EAGA+FI+V+ ST+TSKWFG+ EK +ALF+ A+K++P IIFVDEVDS+L
Sbjct: 1064KTLLAKALATEAGANFISVTGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLL 1243
Query: 626 GQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMV 685
G R EHEA R+++NEFM+ WDGL SK RIL+L ATNRPFDLD+A+IRR RRI V
Sbjct: 1244GARGGAHEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYV 1423
Query: 686 GLPSAENRENILRTLLAKE 704
LP ENR ILR LAKE
Sbjct: 1424DLPDVENRMKILRIFLAKE 1480
Score = 42.7 bits (99), Expect(2) = 1e-84
Identities = 18/38 (47%), Positives = 26/38 (68%)
Frame = +1
Query: 698 RTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAA 735
+ + K ++ + +LA++TEGYSGSDLKNLC AA
Sbjct: 1459 KNISCKRNLNPDFQYDKLASLTEGYSGSDLKNLCVAAA 1572
Score = 28.9 bits (63), Expect = 8.5
Identities = 25/89 (28%), Positives = 39/89 (43%)
Frame = +2
Query: 35 SSSSNALTPDKIENEMLRLVVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSK 94
S + L D+ E+ + VV E V FD+ E + L + ++ E+
Sbjct: 830 SQNLKNLAKDEFESNFVSAVVAPGEIGVKFDDIGAL--EDVKKALQELVILPMRRPELFS 1003
Query: 95 YTRNLAPASRTILLSGPAELYQQVLAKAL 123
+ L P + ILL GP + +LAKAL
Sbjct: 1004HGNLLRPC-KGILLFGPPGTGKTLLAKAL 1087
>TC89903 similar to GP|15810167|gb|AAL06985.1 At1g64110/F22C12_22
{Arabidopsis thaliana}, partial (20%)
Length = 875
Score = 271 bits (693), Expect = 8e-73
Identities = 141/192 (73%), Positives = 165/192 (85%), Gaps = 1/192 (0%)
Frame = +1
Query: 641 KNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTL 700
KNEFM++WDGL S ++ILVL ATNRPFDLDEAIIRRFERRIMVGLPSAENRE IL+TL
Sbjct: 1 KNEFMTHWDGLLSGPNEQILVLGATNRPFDLDEAIIRRFERRIMVGLPSAENREMILKTL 180
Query: 701 LAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD-SKKKKDAEG 759
++KEK H LDFKELATMTEG+SGSDLKNLC TAAYRPVRELIQQE +K+ KKKK+AE
Sbjct: 181 MSKEK-HENLDFKELATMTEGFSGSDLKNLCITAAYRPVRELIQQERKKEMEKKKKEAES 357
Query: 760 QNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEG 819
NS+DA ++ + E E I LRPL+M+D K AK+QVAASFA+EG+ M EL+QWN+LYGEG
Sbjct: 358 VNSEDASNSNDNDEPE--IILRPLSMEDMKQAKNQVAASFASEGSVMAELKQWNELYGEG 531
Query: 820 GSRKKEQLSYFL 831
GSRKK+QL+YFL
Sbjct: 532 GSRKKQQLTYFL 567
>TC86562 similar to GP|21553404|gb|AAM62497.1 26S proteasome regulatory
particle chain RPT6-like protein {Arabidopsis thaliana},
partial (77%)
Length = 1564
Score = 259 bits (662), Expect = 3e-69
Identities = 133/253 (52%), Positives = 178/253 (69%), Gaps = 1/253 (0%)
Frame = +1
Query: 495 VPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGG-LLKPC 553
+ N +E I +VI + I V F IG L+ K +L ELV+LPL+RPDLF G LL P
Sbjct: 334 IQTNSYEDVIACDVINPDHIDVEFDSIGGLETIKQTLFELVILPLQRPDLFSHGKLLGPQ 513
Query: 554 RGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSP 613
+G+LL+GPPGTGKTMLAKAIA E+GA FINV +S + SKWFG+ +K V A+F+LA K+ P
Sbjct: 514 KGVLLYGPPGTGKTMLAKAIAKESGAVFINVRISNLMSKWFGDAQKLVAAVFSLAHKLQP 693
Query: 614 TIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDE 673
+IIF+DEVDS LGQR R +HEA+ +K EFM+ WDG + R++VLAATNRP +LDE
Sbjct: 694 SIIFIDEVDSFLGQR-RSSDHEAVLNMKTEFMALWDGFATDQSARVMVLAATNRPSELDE 870
Query: 674 AIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTT 733
AI+RR + +G P + R +IL+ +L EKV +DF +A + +GY+GSDL +LC
Sbjct: 871 AILRRLPQAFEIGYPDRKERADILKVILKGEKVEDNIDFSYIAGLCKGYTGSDLFDLCKK 1050
Query: 734 AAYRPVRELIQQE 746
AAY P+RE++ E
Sbjct: 1051AAYFPIREILHNE 1089
>TC87563 similar to GP|13027363|dbj|BAB32724. hypothetical protein~similar
to Arabidopsis thaliana chromosome 1 F22C12.12, partial
(14%)
Length = 900
Score = 239 bits (610), Expect(2) = 5e-66
Identities = 130/233 (55%), Positives = 169/233 (71%), Gaps = 1/233 (0%)
Frame = +1
Query: 1 MEQKGMLISA-ALSVGVGVGLGLASGQTMFKPNTYSSSSNALTPDKIENEMLRLVVDGRE 59
MEQK + +SA ++ VG+GVGLGL+SGQ + K + ++ ++I E+ RLVVDG++
Sbjct: 121 MEQKHVFLSALSVGVGLGVGLGLSSGQAVQKWVGGIRDIDEVSGEQIVEELKRLVVDGKD 300
Query: 60 SNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVL 119
S VTFD FPYYLSE+TRVLLTSAAYVHLKH SK+TRNL+PASR ILLSGPAELYQQ+L
Sbjct: 301 SKVTFDEFPYYLSEKTRVLLTSAAYVHLKHLNFSKHTRNLSPASRAILLSGPAELYQQML 480
Query: 120 AKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSETALARLSDLFGSFALFP 179
AKAL HYF++KLLL DVTDFS+K+QSR+G E F RS SE L R+S LFGSF++ P
Sbjct: 481 AKALAHYFDSKLLLLDVTDFSVKLQSRFGIPRKEAYFRRSISEMTLERMSGLFGSFSMMP 660
Query: 180 QREENQGKIHRQSSGSDLRQMEAEGSYSKLRRNASASANISSIGLQSNPTNSA 232
E + + +Q+ +E+ + +LRRNASA++++SS QS TN A
Sbjct: 661 STGETRSTLRQQN-------VESSNNPPRLRRNASAASDLSSTSSQSGSTNPA 798
Score = 31.2 bits (69), Expect(2) = 5e-66
Identities = 13/24 (54%), Positives = 19/24 (79%)
Frame = +2
Query: 242 FDEKILIQTLYKVLLYVSKTYPIV 265
FDEK+ +Q+LYKVL+ +S T I+
Sbjct: 827 FDEKLFVQSLYKVLVSISGTDSII 898
>TC79040 homologue to PIR|F84674|F84674 probable AAA-type ATPase [imported]
- Arabidopsis thaliana, partial (74%)
Length = 1193
Score = 187 bits (476), Expect = 1e-47
Identities = 100/228 (43%), Positives = 148/228 (64%), Gaps = 2/228 (0%)
Frame = +3
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V ++D+ L+ K +LQE V+LP++ P F G +P R LL+GPPGTGK+ LAKA+A
Sbjct: 51 VKWNDVAGLESAKQALQEAVILPVKFPQFFTGKR-RPWRAFLLYGPPGTGKSYLAKAVAT 227
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
EA ++F ++S S + SKW GE EK V LF +A + +P+IIFVDE+DS+ GQR E E
Sbjct: 228 EADSTFFSISSSDLVSKWMGESEKLVSNLFQMARESAPSIIFVDEIDSLCGQRGEGNESE 407
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENREN 695
A R+IK E + G+ ++ ++LVLAATN P+ LD+AI RRF++RI + LP + R++
Sbjct: 408 ASRRIKTELLVQMQGV-GNNDQKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQH 584
Query: 696 ILRTLLAKEKVHG--GLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
+ + L + H D++ LA+ TEG+SGSD+ + PVR+
Sbjct: 585 MFKVHLG-DTPHNLTEKDYEYLASRTEGFSGSDISVCVKDVLFEPVRK 725
>TC85904 similar to PIR|F84674|F84674 probable AAA-type ATPase [imported] -
Arabidopsis thaliana, complete
Length = 1793
Score = 187 bits (475), Expect = 1e-47
Identities = 102/228 (44%), Positives = 146/228 (63%), Gaps = 2/228 (0%)
Frame = +2
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V ++D+ L+ K SLQE V+LP++ P F G +P R LL+GPPGTGK+ LAKA+A
Sbjct: 569 VKWNDVAGLESAKQSLQEAVILPVKFPQFFTGKR-RPWRAFLLYGPPGTGKSYLAKAVAT 745
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
EA ++F +VS S + SKW GE EK V LF +A + +P+IIFVDE+DS+ G R E E
Sbjct: 746 EADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESAPSIIFVDEIDSLCGTRGEGNESE 925
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENREN 695
A R+IK E + G+ ++ ++LVLAATN P+ LD+AI RRF++RI + LP + R++
Sbjct: 926 ASRRIKTELLVQMQGV-GHNDQKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQH 1102
Query: 696 ILRTLLAKEKVHG--GLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
+ + L + H DF+ LA TEG+SGSD+ + PVR+
Sbjct: 1103MFKVHLG-DTPHNLTESDFEHLARKTEGFSGSDIAVCVKDVLFEPVRK 1243
>TC85768 homologue to SP|P54774|CC48_SOYBN Cell division cycle protein 48
homolog (Valosin containing protein homolog) (VCP).
[Soybean], partial (95%)
Length = 2785
Score = 182 bits (463), Expect = 4e-46
Identities = 95/261 (36%), Positives = 159/261 (60%), Gaps = 4/261 (1%)
Frame = +2
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
++ DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIANE
Sbjct: 1529 SWDDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANE 1708
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE-H 634
A+FI++ + + WFGE E NVR +F A +P ++F DE+DS+ QR + VG+
Sbjct: 1709 CQANFISIKGPELLTMWFGESEANVREIFDKARGSAPCVLFFDELDSIATQRGSSVGDAG 1888
Query: 635 EAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAEN 692
A ++ N+ ++ DG+++K + ++ ATNRP +D A++R R ++ I + LP ++
Sbjct: 1889 GAADRVLNQLLTEMDGMSAKK--TVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEDS 2062
Query: 693 RENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSK 752
R I + L K + +D + LA T+G+SG+D+ +C A +RE I+++I+K+ K
Sbjct: 2063 RHQIFKACLRKSPISKDVDIRALAKYTQGFSGADITEICQRACKYAIRENIEKDIEKERK 2242
Query: 753 KKKDAEGQNSQDAQDAKEEVE 773
+ ++ E +D +D E++
Sbjct: 2243 RSENPEAM-EEDIEDEVAEIK 2302
Score = 172 bits (435), Expect = 6e-43
Identities = 95/228 (41%), Positives = 143/228 (62%), Gaps = 2/228 (0%)
Frame = +2
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G + + ++ELV LPLR P LF+ +KP +GILL+GPPG+GKT++A+A+AN
Sbjct: 707 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVAN 886
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E GA F ++ I SK GE E N+R F A K +P+IIF+DE+DS+ +R + E
Sbjct: 887 ETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HGE 1063
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
R+I ++ ++ DGL KS ++V+ ATNRP +D A+ R RF+R I +G+P R
Sbjct: 1064VERRIVSQLLTLMDGL--KSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEVGR 1237
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
+LR K+ +D ++++ T GY G+DL LCT AA + +RE
Sbjct: 1238LEVLRIHTKNMKLAEDVDLEKISKETHGYVGADLAALCTEAALQCIRE 1381
>AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33019
[imported] - Arabidopsis thaliana, partial (34%)
Length = 556
Score = 167 bits (424), Expect = 1e-41
Identities = 83/173 (47%), Positives = 119/173 (67%), Gaps = 6/173 (3%)
Frame = +3
Query: 515 GVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIA 574
GV + D+ L E K L+E V+LPL P+ F+G + +P +G+L+FGPPGTGKT+LAKA+A
Sbjct: 39 GVRWDDVAGLTEAKRLLEEAVVLPLWMPEYFQG-IRRPWKGVLMFGPPGTGKTLLAKAVA 215
Query: 575 NEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEH 634
E G +F NVS +T+ SKW GE E+ VR LF LA +P+ IF+DE+DS+ R GEH
Sbjct: 216 TECGTTFFNVSSATLASKWRGESERMVRCLFDLARAYAPSTIFIDEIDSLCNARGASGEH 395
Query: 635 EAMRKIKNEFMSNWDGLTSK------SEDRILVLAATNRPFDLDEAIIRRFER 681
E+ R++K+E + DG+++ S ++VLAATN P+D+DEA+ RR E+
Sbjct: 396 ESSRRVKSELLVQVDGVSNSATNEDGSRKLVMVLAATNFPWDIDEALRRRLEK 554
>TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AAA-ATPase
subunit RPT4a-like protein {Arabidopsis thaliana},
complete
Length = 1640
Score = 159 bits (402), Expect = 4e-39
Identities = 90/252 (35%), Positives = 150/252 (58%), Gaps = 9/252 (3%)
Frame = +2
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
+++S +G L + L+E + LPL P+LF +KP +G+LL+GPPGTGKT+LA+AIA+
Sbjct: 476 ISYSAVGGLSDQIRELRESIELPLMNPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIAS 655
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVG--- 632
A+F+ V S I K+ GE + +R +F A P IIF+DE+D++ G+R G
Sbjct: 656 NIDANFLKVVSSAIIDKYIGESARLIREMFGYARDHQPCIIFMDEIDAIGGRRFSEGTSA 835
Query: 633 EHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSA 690
+ E R + E ++ DG ++ ++ ATNRP LD A++R R +R+I + LP+
Sbjct: 836 DREIQRTLM-ELLNQLDGFDQLG--KVKMIMATNRPDVLDPALLRPGRLDRKIEIPLPNE 1006
Query: 691 ENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE----LIQQE 746
++R IL+ A HG +D++ + + EG++G+DL+N+CT A +R +I ++
Sbjct: 1007QSRMEILKIHAAGIAKHGEIDYEAVVKLAEGFNGADLRNVCTEAGMSAIRAERDYVIHED 1186
Query: 747 IQKDSKKKKDAE 758
K +K +A+
Sbjct: 1187FMKAVRKLNEAK 1222
>TC85936 homologue to GP|13537115|dbj|BAB40755. AtSUG1 {Arabidopsis
thaliana}, partial (98%)
Length = 1633
Score = 156 bits (394), Expect = 4e-38
Identities = 90/240 (37%), Positives = 147/240 (60%), Gaps = 6/240 (2%)
Frame = +1
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEG-GLLKPCRGILLFGPPGTGKTMLAKAIAN 575
T+ IG LD+ ++E++ LP++ P+LFE G+ +P +G+LL+GPPGTGKT+LA+A+A+
Sbjct: 616 TYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQP-KGVLLYGPPGTGKTLLARAVAH 792
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
+FI VS S + K+ GE + VR LF +A + +P+IIF+DE+DS+ R G
Sbjct: 793 HTDCTFIRVSGSELVQKYIGEGSRMVRELFVMAREHAPSIIFMDEIDSIGSARMESGSGN 972
Query: 636 AMRKIKN---EFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSA 690
+++ E ++ DG ++ ++I VL ATNR LD+A++R R +R+I P+
Sbjct: 973 GDSEVQRTMLELLNQLDGF--EASNKIKVLMATNRIDILDQALLRPGRIDRKIEFPNPNE 1146
Query: 691 ENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD 750
E+R +IL+ + + G+D K++A G SG++LK +CT A +RE Q+D
Sbjct: 1147ESRRDILKIHSRRMNLMRGIDLKKIAEKMNGASGAELKAVCTEAGMFALRERRVHVTQED 1326
>TC85745 homologue to GP|17297987|dbj|BAB78491. 26S proteasome regulatory
particle triple-A ATPase subunit2b {Oryza sativa
(japonica cultivar-group), partial (98%)
Length = 1696
Score = 154 bits (390), Expect = 1e-37
Identities = 89/245 (36%), Positives = 142/245 (57%), Gaps = 5/245 (2%)
Frame = +3
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
+++DIG LD ++E V LPL P+L+E +KP +G++L+G PGTGKT+LAKA+AN
Sbjct: 645 SYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANS 824
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRV---GE 633
A+F+ V S + K+ G+ K VR LF +A +SP I+F+DE+D++ +R GE
Sbjct: 825 TSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPAIVFIDEIDAVGTKRYDAHSGGE 1004
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
E R + E ++ DG S+ + + V+ ATNR LD A++R R +R+I LP +
Sbjct: 1005REIQRTML-ELLNQLDGFDSRGD--VKVILATNRIESLDPALLRPGRIDRKIEFPLPDIK 1175
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
R I + ++ + ++ +E + +SG+D+K +CT A +RE + D
Sbjct: 1176TRRRIFQIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRERRMKVTHPDF 1355
Query: 752 KKKKD 756
KK KD
Sbjct: 1356KKAKD 1370
>TC85744 homologue to PIR|T08959|T08959 proteinase homolog F19B15.70 -
Arabidopsis thaliana, complete
Length = 1629
Score = 154 bits (389), Expect = 1e-37
Identities = 89/245 (36%), Positives = 142/245 (57%), Gaps = 5/245 (2%)
Frame = +2
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
+++DIG LD ++E V LPL P+L+E +KP +G++L+G PGTGKT+LAKA+AN
Sbjct: 647 SYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANS 826
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRV---GE 633
A+F+ V S + K+ G+ K VR LF +A +SP+I+F+DE+D++ +R GE
Sbjct: 827 TSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGGE 1006
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
E R + E ++ DG S+ + + V+ ATNR LD A++R R +R+I LP +
Sbjct: 1007REIQRTML-ELLNQLDGFDSRGD--VKVILATNRIESLDPALLRPGRIDRKIEFPLPDIK 1177
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
R I ++ + ++ +E + +SG+D+K +CT A +RE + D
Sbjct: 1178TRRRIFTIHTSRMTLADDVNLEEFVMTKDEFSGADIKAICTEAGLLALRERRMKVTHADF 1357
Query: 752 KKKKD 756
KK KD
Sbjct: 1358KKAKD 1372
>TC87818 similar to GP|20197082|gb|AAC26698.2 putative katanin {Arabidopsis
thaliana}, partial (62%)
Length = 984
Score = 152 bits (383), Expect = 7e-37
Identities = 80/186 (43%), Positives = 118/186 (63%), Gaps = 1/186 (0%)
Frame = +2
Query: 561 PPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDE 620
P G + MLAKA+A E +F N+S S++ SKW G+ EK V+ LF LA +P IF+DE
Sbjct: 5 PQGLERPMLAKAVATECNTTFFNISASSMFSKWRGDSEKLVKVLFELARHHAPATIFLDE 184
Query: 621 VDSMLGQRTRV-GEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRF 679
+D+++ QR EHEA R++K E + DGL +++++ + VLAATN P++LD A++RR
Sbjct: 185 IDAIISQRGEGRSEHEASRRLKTELLIQMDGL-ARTDELVFVLAATNLPWELDAAMLRRL 361
Query: 680 ERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPV 739
E+RI+V LP E R + LL + + + L TEGYSGSD++ LC A +P+
Sbjct: 362 EKRILVPLPEPEARRAMFEELLPLQPDEEPMPYDLLVDRTEGYSGSDIRLLCKETAMQPL 541
Query: 740 RELIQQ 745
R L+ Q
Sbjct: 542 RRLMTQ 559
>TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog chloroplast
precursor (EC 3.4.24.-). [Alfalfa] {Medicago sativa},
complete
Length = 2473
Score = 147 bits (371), Expect = 2e-35
Identities = 92/242 (38%), Positives = 137/242 (56%), Gaps = 4/242 (1%)
Frame = +3
Query: 513 EIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKA 572
E GVTF+D+ D+ K LQE+V L+ PD + K +G LL GPPGTGKT+LA+A
Sbjct: 990 ETGVTFADVAGADQAKLELQEVVDF-LKNPDKYTALGAKIPKGCLLVGPPGTGKTLLARA 1166
Query: 573 IANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTR-- 630
+A EAG F + + S + G VR LF A +P I+F+DE+D++ QR
Sbjct: 1167 VAGEAGTPFFSCAASEFVELFVGVGASRVRDLFEKAKSKAPCIVFIDEIDAVGRQRGAGL 1346
Query: 631 VGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLP 688
G ++ + N+ ++ DG + S ++VLAATNRP LD A++R RF+R++ V P
Sbjct: 1347 GGGNDEREQTINQLLTEMDGFSGNS--GVIVLAATNRPDVLDSALLRPGRFDRQVTVDRP 1520
Query: 689 SAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQ 748
R IL+ + + +DF ++A T G++G+DL+NL AA R + +EI
Sbjct: 1521 DVAGRVKILQVHSRGKALAKDVDFDKIARRTPGFTGADLQNLMNEAAILAARRDL-KEIS 1697
Query: 749 KD 750
KD
Sbjct: 1698 KD 1703
>TC77012 homologue to SP|P54776|PRSA_LYCES 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1)
(Mg(2+), complete
Length = 1793
Score = 144 bits (363), Expect = 1e-34
Identities = 88/267 (32%), Positives = 151/267 (55%), Gaps = 4/267 (1%)
Frame = +1
Query: 518 FSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEA 577
++DIG L++ L E ++LP+ + F+ ++P +G+LL+GPPGTGKT++A+A A +
Sbjct: 637 YNDIGGLEKQIQELVEAIVLPMTHKERFQKLGIRPPKGVLLYGPPGTGKTLMARACAAQT 816
Query: 578 GASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR--TRVGEHE 635
A+F+ ++ + + G+ K VR F LA + SP IIF+DE+D++ +R + V
Sbjct: 817 NATFLKLAGPQLVQMFIGDGAKLVRDAFQLAKEKSPCIIFIDEIDAIGTKRFDSEVSGDR 996
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
+++ E ++ DG + S+DRI V+AATNR LD A++R R +R+I P+ E R
Sbjct: 997 EVQRTMLELLNQLDGFS--SDDRIKVIAATNRADILDPALMRSGRLDRKIEFPHPTEEAR 1170
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKK 753
IL+ K VH ++F+ELA T+ ++G+ LK +C A +R
Sbjct: 1171ARILQIHSRKMNVHPDVNFEELARSTDDFNGAQLKAVCVEAGMLALR------------- 1311
Query: 754 KKDAEGQNSQDAQDAKEEVEQERVITL 780
+DA N +D + +V+ ++ +L
Sbjct: 1312-RDATEVNHEDFNEGIIQVQAKKKASL 1389
>BF647954 homologue to GP|11094192|db 26S proteasome regulatory particle
triple-A ATPase subunit4 {Oryza sativa (japonica
cultivar-group)}, partial (53%)
Length = 649
Score = 143 bits (360), Expect = 3e-34
Identities = 80/214 (37%), Positives = 128/214 (59%), Gaps = 5/214 (2%)
Frame = +2
Query: 542 PDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNV 601
P+LF +KP +G+LL+GPPGTGKT+LA+AIA+ A+F+ V S I K+ GE + +
Sbjct: 8 PELFLRVGIKPPKGVLLYGPPGTGKTLLARAIASNIDANFLKVVSSAIIDKYIGESSRLI 187
Query: 602 RALFTLAAKVSPTIIFVDEVDSMLGQRTRVG---EHEAMRKIKNEFMSNWDGLTSKSEDR 658
R +F A P IIF+DE+D++ G+R G + E R + E ++ DG +
Sbjct: 188 REMFGYARDHQPCIIFMDEIDAIGGRRFSEGTSADREIQRTLM-ELLNQLDGFDQLG--K 358
Query: 659 ILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELA 716
+ ++ ATNRP LD A++R R +R+I + LP+ ++R IL+ A HG +D++ +
Sbjct: 359 VKIIMATNRPDVLDPALLRPGRLDRKIEIPLPNEQSRMEILKIHAAGIAKHGEIDYEAVV 538
Query: 717 TMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD 750
+ EG++G+DL+N+CT A +R I +D
Sbjct: 539 KLAEGFNGADLRNICTEAGMSAIRAERDYVIHED 640
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,092,846
Number of Sequences: 36976
Number of extensions: 234378
Number of successful extensions: 1191
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 1086
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1119
length of query: 831
length of database: 9,014,727
effective HSP length: 104
effective length of query: 727
effective length of database: 5,169,223
effective search space: 3758025121
effective search space used: 3758025121
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140913.6


