

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.4 - phase: 0
(374 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
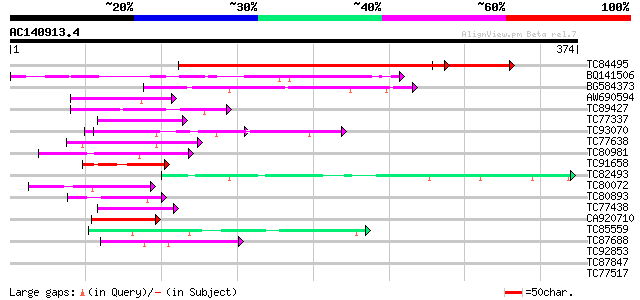
Score E
Sequences producing significant alignments: (bits) Value
TC84495 similar to GP|13027361|dbj|BAB32722. hypothetical protei... 351 3e-97
BQ141506 53 2e-07
BG584373 similar to GP|4056491|gb|A unknown protein {Arabidopsis... 51 8e-07
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 49 2e-06
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 48 7e-06
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 47 9e-06
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 47 1e-05
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 47 2e-05
TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|... 44 1e-04
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 43 2e-04
TC82493 weakly similar to GP|4056491|gb|AAC98057.1| unknown prot... 43 2e-04
TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Ar... 43 2e-04
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 43 2e-04
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 42 4e-04
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 41 7e-04
TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC ... 41 7e-04
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 41 7e-04
TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Ar... 40 0.001
TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-... 40 0.001
TC77517 similar to PIR|T51159|T51159 HMG protein [imported] - Ar... 40 0.001
>TC84495 similar to GP|13027361|dbj|BAB32722. hypothetical protein~similar
to Arabidopsis thaliana chromosome 1 F22C12.16, partial
(15%)
Length = 675
Score = 351 bits (900), Expect = 3e-97
Identities = 179/179 (100%), Positives = 179/179 (100%)
Frame = +2
Query: 112 NNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPNNKPQFTASLLKSAT 171
NNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPNNKPQFTASLLKSAT
Sbjct: 8 NNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPNNKPQFTASLLKSAT 187
Query: 172 KFRVFMSGLKKPKTDSVLEKKQHKPERIKFKVEEVPIVSLFTRDNSSKNSNKSQTNQNQK 231
KFRVFMSGLKKPKTDSVLEKKQHKPERIKFKVEEVPIVSLFTRDNSSKNSNKSQTNQNQK
Sbjct: 188 KFRVFMSGLKKPKTDSVLEKKQHKPERIKFKVEEVPIVSLFTRDNSSKNSNKSQTNQNQK 367
Query: 232 VEHESSLSPLEEKRFSKEIVMHKYLKMVKPLYIRVSRRYSHGDKSSPSEKVEAETATEG 290
VEHESSLSPLEEKRFSKEIVMHKYLKMVKPLYIRVSRRYSHGDKSSPSEKVEAETATEG
Sbjct: 368 VEHESSLSPLEEKRFSKEIVMHKYLKMVKPLYIRVSRRYSHGDKSSPSEKVEAETATEG 544
Score = 90.1 bits (222), Expect = 1e-18
Identities = 44/54 (81%), Positives = 50/54 (92%)
Frame = +3
Query: 280 EKVEAETATEGETVAENEENNAKTQSQKQGNNIPAGLRVVYKHLGKSRSASSAV 333
++++ + +GETVAENEENNAKTQSQKQGNNIPAGLRVVYKHLGKSRSASSAV
Sbjct: 513 KRLKQKQRRKGETVAENEENNAKTQSQKQGNNIPAGLRVVYKHLGKSRSASSAV 674
>BQ141506
Length = 599
Score = 53.1 bits (126), Expect = 2e-07
Identities = 66/272 (24%), Positives = 110/272 (40%), Gaps = 12/272 (4%)
Frame = +1
Query: 1 MEAFSLLKYWRGGGAVVGLSPSSDSTTTTNVNTTTILTTAESSDDNDDEGPFFDLEFTAP 60
M+A + LK+W+ ++TTT+N+ L E+ ++D++ FFDLE T
Sbjct: 25 MDALNFLKFWK------------NNTTTSNITVPPHLVV-ETDSESDEDDSFFDLELTM- 162
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDANL 120
DD +T++E NN L
Sbjct: 163 --------------------------------SDDQHVKTKTE--------NNVPQKTTL 222
Query: 121 SLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPNNKPQFTASLLKSATKFRVF---- 176
+SP+E + K K++ + EP +KPQ SLL+SA FR+F
Sbjct: 223 PMSPNEPIS-KRKVLPI-----------------EPISKPQSPISLLRSAPSFRIFTFRK 348
Query: 177 --MSGLKKP----KTD-SVLEKKQHKPERIKFKVE-EVPIVSLFTRDNSSKNSNKSQTNQ 228
+G +K KT+ +KK+ K +K +E + + +RDNS+++ NQ
Sbjct: 349 QRRTGSEKTEEFRKTECEKNQKKETKVFSVKLNIEHDFHSTPMLSRDNSTRSFGSKIRNQ 528
Query: 229 NQKVEHESSLSPLEEKRFSKEIVMHKYLKMVK 260
+ +S +RF + KYLK++K
Sbjct: 529 GTEEPKTERVS----RRF-----LXKYLKLIK 597
>BG584373 similar to GP|4056491|gb|A unknown protein {Arabidopsis thaliana},
partial (11%)
Length = 767
Score = 50.8 bits (120), Expect = 8e-07
Identities = 54/207 (26%), Positives = 98/207 (47%), Gaps = 26/207 (12%)
Frame = +1
Query: 89 SDDDDDDDSGETESEFKITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFI----- 143
S D S + S+F+ T+S S + S+ +L P+++LF+KG+L+ L S I
Sbjct: 91 SSPSHDSFSSSSSSDFEFTISISPRKSSN---TLCPADELFYKGQLLPLHLSPRISMVRT 261
Query: 144 -LNNNNNNSSSSEPNNKPQFTASLLKSATKFRVFMSGLKKPKTDSVLEKKQHKPERIKFK 202
L ++++ SS++ + P+ + S F+S L DS + + +
Sbjct: 262 LLLSSSSTSSATSSSTAPRDSTGSSASNDSTTSFISDLAL-FPDSCNSSSRPSSVTEEDE 438
Query: 203 VEEVPIVSLFTRDNSS-KNSNK----------------SQTNQNQKVEHESSLSPLEEKR 245
++ + S TR NS K +NK ++T + + V + + S ++ KR
Sbjct: 439 LKRLHNSSNSTRSNSQLKKTNKYFSLSRFSSVFRKETVAKTQEGETVANSINNSSVKMKR 618
Query: 246 FS---KEIVMHKYLKMVKPLYIRVSRR 269
S KE++ KY K VKPLY ++S++
Sbjct: 619 MSVTAKEVI-RKYFKKVKPLYEKLSQK 696
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 49.3 bits (116), Expect = 2e-06
Identities = 24/77 (31%), Positives = 43/77 (55%), Gaps = 7/77 (9%)
Frame = +3
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDD-------DEDSDDDD 93
E ++ +DEG + E EEEED +E + +++EEE + DD +E+ +++D
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEED 224
Query: 94 DDDSGETESEFKITLSP 110
DD+ GE + +I+ P
Sbjct: 225 DDEEGEEDEIDRISTLP 275
Score = 38.1 bits (87), Expect = 0.006
Identities = 16/56 (28%), Positives = 31/56 (54%)
Frame = +3
Query: 62 EEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
EEED E +++ EEE+DE D+E+ +++++++ E + E + E D
Sbjct: 57 EEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEED 224
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 47.8 bits (112), Expect = 7e-06
Identities = 34/109 (31%), Positives = 52/109 (47%), Gaps = 3/109 (2%)
Frame = +1
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGET 100
+ DD +DE D +F+ E++ED E + D+DD++D DDDDD+D GE
Sbjct: 328 DDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPV-PNGAGGSDDDDEDDDDDDDDNDDGED 504
Query: 101 ESEFKITLSPSNNERSDANLSLSPSED---LFFKGKLVQLKSSSFILNN 146
E E +E D + PS+ + GKL L + FI ++
Sbjct: 505 EDE---------DEEEDDDEDQPPSKKKK*VNIVGKLFFLPFAHFIFSS 624
Score = 36.6 bits (83), Expect = 0.016
Identities = 16/39 (41%), Positives = 25/39 (64%), Gaps = 4/39 (10%)
Frame = +1
Query: 69 ETNHKNQQQEEEKDEDDDEDSDDDDDDD----SGETESE 103
E N + E++ DEDDD+D +D+DDD+ SG+ + E
Sbjct: 283 EENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDE 399
Score = 34.3 bits (77), Expect = 0.080
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Frame = +1
Query: 61 EEEEDGFEETNHKNQQQEEE-KDEDDDEDSDDDDDDDSGETESE 103
EE +DG E + ++ +++ DEDDD D D D+D + + E
Sbjct: 283 EENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPE 414
Score = 34.3 bits (77), Expect = 0.080
Identities = 13/42 (30%), Positives = 25/42 (58%), Gaps = 2/42 (4%)
Frame = +1
Query: 65 DGFEETNHKNQQQEEEKDEDDDEDSDDDDD--DDSGETESEF 104
D + NH ++ ++ + +DD+D DDDDD D+ + + +F
Sbjct: 253 DQLSKGNHTCEENKDGSETEDDDDEDDDDDVNDEDDDNDEDF 378
Score = 33.9 bits (76), Expect = 0.10
Identities = 15/45 (33%), Positives = 23/45 (50%)
Frame = +1
Query: 59 APEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
+P + ++ + N EE KD + ED DD+DDDD E +
Sbjct: 226 SPMDGRFPLDQLSKGNHTCEENKDGSETEDDDDEDDDDDVNDEDD 360
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 47.4 bits (111), Expect = 9e-06
Identities = 21/60 (35%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Frame = +3
Query: 59 APEEEEDGFEETNHKNQQQE-EEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSD 117
APE G +E + ++ +++DEDDDED DDDD+++ GE + E + + E D
Sbjct: 555 APEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEED 734
Score = 37.7 bits (86), Expect = 0.007
Identities = 17/57 (29%), Positives = 35/57 (60%)
Frame = +3
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDS 97
E+ ++ DDE D + +E+ED +E + +EE DE+D+E+ ++D+D+++
Sbjct: 585 ENGEEEDDEDG--DDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEEA 749
Score = 33.1 bits (74), Expect = 0.18
Identities = 19/67 (28%), Positives = 28/67 (41%), Gaps = 2/67 (2%)
Frame = +3
Query: 60 PEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSP--SNNERSD 117
P + D ++ K E E DD+++ DDD D GE E E NN +
Sbjct: 360 PTTKGDSKTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNK 539
Query: 118 ANLSLSP 124
+N +P
Sbjct: 540 SNSKKAP 560
Score = 33.1 bits (74), Expect = 0.18
Identities = 21/80 (26%), Positives = 40/80 (49%), Gaps = 4/80 (5%)
Frame = +3
Query: 40 AESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQ----EEEKDEDDDEDSDDDDDD 95
A +D+N +E E++EDG ++ ++ + EEE E+D+E+ D++D+
Sbjct: 570 AGGADENGEE-----------EDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDN 716
Query: 96 DSGETESEFKITLSPSNNER 115
+ E E E + L P +
Sbjct: 717 EE-EEEDEDEEALQPPKKRK 773
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 47.0 bits (110), Expect = 1e-05
Identities = 46/176 (26%), Positives = 80/176 (45%), Gaps = 9/176 (5%)
Frame = +1
Query: 56 EFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNER 115
+F +P + +D EE + ++ + E+E +++DD+D DDDDD+D + E
Sbjct: 202 KFLSPYDSDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDD----------DGFAEP 351
Query: 116 SDANLSLSPSEDLFFKGKLV----QLKSSSFILNNNNNNSSSSEPNNKPQFTASLLK-SA 170
+D N ++D K K V Q K + + NN S P K Q ASL + ++
Sbjct: 352 TDLN-----AKDKRLKSKTVPDRQQEKEKEKGVRSLNNGQSKRLP--KSQRIASLQENNS 510
Query: 171 TKFRVFMSGLKKPK-TDSVLEKKQHKP---ERIKFKVEEVPIVSLFTRDNSSKNSN 222
KFR K P+ ++ +L ++ P F ++E + ++ R S N
Sbjct: 511 AKFRRNSMEKKYPELSEEILLDEKWLPLLDYLSTFGIKESQFIQIYERHMSXFQIN 678
Score = 45.4 bits (106), Expect = 3e-05
Identities = 29/115 (25%), Positives = 50/115 (43%), Gaps = 6/115 (5%)
Frame = +1
Query: 50 GPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDD------DSGETESE 103
G F + E EE FE+ +++ ++E+ D+DDD+D +DDDDD D +
Sbjct: 199 GKFLSPYDSDDEIEEMDFEDDEDEDEDEDEDDDDDDDDDDEDDDDDGFAEPTDLNAKDKR 378
Query: 104 FKITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPNN 158
K P + + + + G+ +L S I + NNS+ N+
Sbjct: 379 LKSKTVPDRQQEKEKEKGVRSLNN----GQSKRLPKSQRIASLQENNSAKFRRNS 531
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 46.6 bits (109), Expect = 2e-05
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 9/99 (9%)
Frame = +1
Query: 38 TTAESSDDN--DDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDD 95
+T +SS+D +DEG + E + E +DG E + +++ E EK ++ +E++ D D+
Sbjct: 619 STEKSSEDTKTEDEGKKTEDEGSNTENNKDGEEASTKESESDESEKKDESEENNKSDSDE 798
Query: 96 ------DSGE-TESEFKITLSPSNNERSDANLSLSPSED 127
DS E T+S + + S N+ SD N S ++D
Sbjct: 799 SEKKSSDSNETTDSNVEEKVEQSQNKESDENASEKNTDD 915
>TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|AI994480
gb|T44186 gb|Z30806 come from this gene. {Arabidopsis
thaliana}, partial (37%)
Length = 699
Score = 43.5 bits (101), Expect = 1e-04
Identities = 31/114 (27%), Positives = 52/114 (45%), Gaps = 12/114 (10%)
Frame = +3
Query: 20 SPSSDSTTTTNVNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEE 79
+P STTT + T T SSD + P E +++ED +E N + +
Sbjct: 144 NPKLTSTTTVDQETELPTTVNSSSDAVPNNEP----ENNTDDDDEDDEDEENSDEEPVVD 311
Query: 80 EKDED------------DDEDSDDDDDDDSGETESEFKITLSPSNNERSDANLS 121
K + ++ED DDDD DDS +++ + +S S+++ SD L+
Sbjct: 312 RKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLA 473
Score = 27.7 bits (60), Expect = 7.5
Identities = 16/76 (21%), Positives = 30/76 (39%)
Frame = +3
Query: 28 TTNVNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDE 87
T N T T + + P + + T+ + E N + + + +
Sbjct: 66 TNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPEN 245
Query: 88 DSDDDDDDDSGETESE 103
++DDDD+DD E S+
Sbjct: 246 NTDDDDEDDEDEENSD 293
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 43.1 bits (100), Expect = 2e-04
Identities = 21/57 (36%), Positives = 37/57 (64%)
Frame = +2
Query: 49 EGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFK 105
E F D+E E++EDG EE + +++ D+DDDE+ +DD+++D G+ +S+ K
Sbjct: 59 ESDFDDIE--VDEDDEDGDEEDD------DDDDDDDDDEEDEDDEEEDEGKGKSKSK 205
Score = 38.1 bits (87), Expect = 0.006
Identities = 13/43 (30%), Positives = 26/43 (60%)
Frame = +2
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
E E F++ ++ ++++DDD+D DDDD++D + E +
Sbjct: 50 EAGESDFDDIEVDEDDEDGDEEDDDDDDDDDDDEEDEDDEEED 178
Score = 37.4 bits (85), Expect = 0.009
Identities = 16/59 (27%), Positives = 30/59 (50%)
Frame = +2
Query: 69 ETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDANLSLSPSED 127
E++ + + +E+ ++ D+ED DDDDDDD E + + + +S P +D
Sbjct: 59 ESDFDDIEVDEDDEDGDEEDDDDDDDDDDDEEDEDDEEEDEGKGKSKSKRGSKAKPGKD 235
>TC82493 weakly similar to GP|4056491|gb|AAC98057.1| unknown protein
{Arabidopsis thaliana}, partial (35%)
Length = 1158
Score = 43.1 bits (100), Expect = 2e-04
Identities = 66/303 (21%), Positives = 119/303 (38%), Gaps = 30/303 (9%)
Frame = +1
Query: 101 ESEFKITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFI-----LNNNNNNSSSSE 155
E EF+++ S SN + S +P++DLF+KGKL+ L + L N N ++
Sbjct: 187 EFEFQMSSSISNEKNSKT----TPADDLFYKGKLLPLHLPRRLQMVQKLLENTNLEDNTF 354
Query: 156 PNNKPQFTASLLKSATKFRVFMSGLKKPKTDSVLEKKQHKPERIKFKVEEVPIVSLFTRD 215
P P F + T +SG + + S + ++ E F V ++P
Sbjct: 355 PFTPPTFAS----YTTLETCNISGSESCRVSSDVSPDEYSFEMNGFVVRDLP-------- 498
Query: 216 NSSKNSNKSQTNQNQKVEHESSLSPLEEKRFSKEIVMHKYLKMVKPLYIRVSRRYSHGDK 275
NKS ++ + +++ + LK K + + S +K
Sbjct: 499 -----KNKSWP---------------KKLKMMNQLLFGQKLKASKAYLKSLFNKTSCSEK 618
Query: 276 ---SSPSEKVEAETATEGETVAENEENNAKTQSQKQG----NNIPAGLRVVYKHLGKSRS 328
S P+ KV ++ + EEN + + N+ + VV +H G ++
Sbjct: 619 ACASDPNNKVGIKSKNKNSFEVFCEENKKQVKRDMVEDDFVNHRKSFSGVVQRHCGSNKV 798
Query: 329 ASSAVAASPPVLVSS---------------KRRDDSLLQQQDGIQGAILHCKRS---FNA 370
+S + ++S SS KR + + + ++GAI HCK+S FN+
Sbjct: 799 SSLSTSSSGSSSNSSSFSFSSSGNYDLQLFKRSISANYEVEGSVEGAIAHCKQSQQHFNS 978
Query: 371 SRG 373
G
Sbjct: 979 KNG 987
>TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 746
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/93 (30%), Positives = 40/93 (42%), Gaps = 9/93 (9%)
Frame = +1
Query: 13 GGAVVGLSPSSDSTTTTNVNTTTILTTAESSDDNDDEGPFF---------DLEFTAPEEE 63
GG D+TTT+ + S++D DDE F D+ + E +
Sbjct: 277 GGGSKSYQKKKDATTTSRRDRYNF-----SAEDMDDEIDIFHKQRDIVPLDINADSAESD 441
Query: 64 EDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDD 96
ED + +E DED DE+ DDDD+DD
Sbjct: 442 EDDEMPIFNDKDIDNDETDEDGDEEEDDDDEDD 540
Score = 32.0 bits (71), Expect = 0.40
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Frame = +1
Query: 42 SSDDNDDEGPFF-DLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGE 99
+ D DDE P F D + E +EDG EEE D+D+D++ D + G+
Sbjct: 430 AESDEDDEMPIFNDKDIDNDETDEDG----------DEEEDDDDEDDEYDGKEKGFIGQ 576
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 42.7 bits (99), Expect = 2e-04
Identities = 23/72 (31%), Positives = 37/72 (50%), Gaps = 7/72 (9%)
Frame = +3
Query: 39 TAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDS-------DD 91
T + DD+DD+ ++E+D EE ++ + EEE D +DD ++ D
Sbjct: 321 TEDDEDDDDDDDV---------QDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDG 473
Query: 92 DDDDDSGETESE 103
+DDDD G+ E E
Sbjct: 474 EDDDDDGDEEDE 509
Score = 38.5 bits (88), Expect = 0.004
Identities = 15/30 (50%), Positives = 20/30 (66%)
Frame = +3
Query: 72 HKNQQQEEEKDEDDDEDSDDDDDDDSGETE 101
+K + + E DEDDD+D D D+DD GE E
Sbjct: 300 YKENKSDTEDDEDDDDDDDVQDEDDDGEEE 389
Score = 32.3 bits (72), Expect = 0.30
Identities = 14/42 (33%), Positives = 23/42 (54%)
Frame = +3
Query: 60 PEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETE 101
P E G + +N+ E+ ++DDD+D D+DDD E +
Sbjct: 267 PLEHLLGRKPAYKENKSDTEDDEDDDDDDDVQDEDDDGEEED 392
Score = 30.4 bits (67), Expect = 1.2
Identities = 16/52 (30%), Positives = 28/52 (53%)
Frame = +3
Query: 40 AESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDD 91
++ +D+DD+G E+EEDG E+E+DE+D+E+ D+
Sbjct: 462 SDDGEDDDDDGD--------EEDEEDG-----------EDEEDEEDEEEDDE 560
Score = 30.0 bits (66), Expect = 1.5
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 6/39 (15%)
Frame = +3
Query: 71 NHKNQQQEEEKDEDDDEDSDDDDDD------DSGETESE 103
N + + +E+ D+DDD +DDD + D GE E +
Sbjct: 309 NKSDTEDDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGD 425
Score = 28.9 bits (63), Expect = 3.4
Identities = 10/25 (40%), Positives = 16/25 (64%)
Frame = +3
Query: 79 EEKDEDDDEDSDDDDDDDSGETESE 103
+E D ++D DDDDDDD + + +
Sbjct: 303 KENKSDTEDDEDDDDDDDVQDEDDD 377
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 - garden
pea, partial (93%)
Length = 1607
Score = 42.0 bits (97), Expect = 4e-04
Identities = 19/53 (35%), Positives = 30/53 (55%)
Frame = +2
Query: 59 APEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPS 111
A EE ++ + E+ DED+DED D+DDD+D ET+++ K + S
Sbjct: 1010 AQGEEFGDLDDEDDDEDDDAEDDDEDEDEDEDEDDDEDEEETKTKKKSSAKKS 1168
Score = 40.0 bits (92), Expect = 0.001
Identities = 20/64 (31%), Positives = 34/64 (52%)
Frame = +2
Query: 57 FTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERS 116
FT + + F + + ++ ++++ EDDDED D+D+D+D E E E K S +
Sbjct: 995 FTGEAAQGEEFGDLDDEDDDEDDDA-EDDDEDEDEDEDEDDDEDEEETKTKKKSSAKKSG 1171
Query: 117 DANL 120
A L
Sbjct: 1172 IAQL 1183
Score = 34.3 bits (77), Expect = 0.080
Identities = 14/44 (31%), Positives = 25/44 (56%)
Frame = +2
Query: 78 EEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDANLS 121
++E D++DD+ DDD+D+D E E + + ++S A S
Sbjct: 1037 DDEDDDEDDDAEDDDEDEDEDEDEDDDEDEEETKTKKKSSAKKS 1168
Score = 27.7 bits (60), Expect = 7.5
Identities = 12/50 (24%), Positives = 29/50 (58%)
Frame = +2
Query: 56 EFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFK 105
EF ++E+D ++ + + E+E +++DD D+D+++ + +S K
Sbjct: 1022 EFGDLDDEDDDEDDDAEDDDEDEDEDEDEDD---DEDEEETKTKKKSSAK 1162
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 41.2 bits (95), Expect = 7e-04
Identities = 18/46 (39%), Positives = 30/46 (65%), Gaps = 1/46 (2%)
Frame = -2
Query: 55 LEFTAPEEEEDGFEETNHK-NQQQEEEKDEDDDEDSDDDDDDDSGE 99
+E P + G+ E + +++EEEKDED+DE+ D+D+D+D E
Sbjct: 425 VESLVPTPWKSGYIEAEAEAEEEEEEEKDEDEDENEDEDEDEDEDE 288
Score = 33.9 bits (76), Expect = 0.10
Identities = 13/37 (35%), Positives = 24/37 (64%)
Frame = -2
Query: 62 EEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSG 98
E E EE + + ++E+++ED+DED D+D++ G
Sbjct: 383 EAEAEAEEEEEEEKDEDEDENEDEDEDEDEDEEVAGG 273
Score = 32.7 bits (73), Expect = 0.23
Identities = 12/35 (34%), Positives = 24/35 (68%)
Frame = -2
Query: 52 FFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDD 86
+ + E A EEEE+ +E +N+ ++E++DED++
Sbjct: 389 YIEAEAEAEEEEEEEKDEDEDENEDEDEDEDEDEE 285
>TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC 6.4.1.2) -
potato chloroplast, partial (66%)
Length = 5011
Score = 41.2 bits (95), Expect = 7e-04
Identities = 47/204 (23%), Positives = 80/204 (39%), Gaps = 18/204 (8%)
Frame = +3
Query: 53 FDLEFTAPEEEEDGFEETNHKNQQQEE---------EKDEDDDEDSDDDDDDDSGETESE 103
F++ F ++E + K Q+ + E D D DSDD+D DD +T +
Sbjct: 2685 FEVRFETDSDKEKDSGNDDDKLQKASDILEPVNYSAENYSDSDNDSDDNDPDDDYDTLQK 2864
Query: 104 FKITLSPSNNERSD-------ANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEP 156
TL P N+ +D A+ L P D S ++ + EP
Sbjct: 2865 GTDTLEPENDNDTDDYDKIQRAHYILEPEND------------SDTEPDDEPDYEPDDEP 3008
Query: 157 NNKPQFTASLLKSATKFRVFMSGLKKPKTDSVLEKKQHKPERIKFKVEEVPIVSLFTRDN 216
+++P L K+ S + +P+ S EK Q+ E + E P S +++
Sbjct: 3009 DDEPDDDDKLQKA--------SDILEPENPSDSEKVQNSSEIESTGIMENPSDSEKVQNS 3164
Query: 217 SSKNSNKSQTN--QNQKVEHESSL 238
S S N ++KV++ S +
Sbjct: 3165 SEIESTGIMENPSDSEKVQNSSEI 3236
Score = 40.4 bits (93), Expect = 0.001
Identities = 30/114 (26%), Positives = 48/114 (41%), Gaps = 17/114 (14%)
Frame = +3
Query: 19 LSPSSDSTTTTNVNTTTILTTAESSDDNDDEGPFFDL----EFTAPEEEEDG-------- 66
L +SD N + + SDDND + + L + PE + D
Sbjct: 2748 LQKASDILEPVNYSAENYSDSDNDSDDNDPDDDYDTLQKGTDTLEPENDNDTDDYDKIQR 2927
Query: 67 ----FEETNHKNQQQEEEKD-EDDDEDSDDDDDDDSGETESEFKITLSPSNNER 115
E N + + ++E D E DDE D+ DDDD + S+ +PS++E+
Sbjct: 2928 AHYILEPENDSDTEPDDEPDYEPDDEPDDEPDDDDKLQKASDILEPENPSDSEK 3089
Score = 32.3 bits (72), Expect = 0.30
Identities = 52/261 (19%), Positives = 102/261 (38%), Gaps = 36/261 (13%)
Frame = +3
Query: 24 DSTTTTNVNTTTILTTAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDE 83
D T T T+ E+ +D DD + E + E + + + ++E D+
Sbjct: 2841 DDYDTLQKGTDTL--EPENDNDTDDYDKIQRAHYILEPENDSDTEPDDEPDYEPDDEPDD 3014
Query: 84 DDDED---------------SDDDDDDDSGETESEFKITLSPSNNERSDANLSLSPSEDL 128
+ D+D SD + +S E ES I +PS++E+ + + + +
Sbjct: 3015 EPDDDDKLQKASDILEPENPSDSEKVQNSSEIEST-GIMENPSDSEKVQNSSEIESTGIM 3191
Query: 129 FFKGKLVQLKSSSFILN-------------------NNNNNSSSSEPNNKPQFTASLLKS 169
++++SS I + NN N ++ LK
Sbjct: 3192 ENPSDSEKVQNSSEIESTGIIRKDFSHLWVACDSCYGNNYKRFFKSKMNICEYCGCHLKM 3371
Query: 170 ATKFRVFMSGLKKPKTDSVLEKKQHKPERIKFKVEEVPI-VSLFTRDNSSKNSNKSQTNQ 228
++ R+ + L P T + +++ + I+F E+ P L D S+NS + + ++
Sbjct: 3372 SSSDRIEL--LIDPGTWNPMDEDMFPVDPIEFNSEDEPSEKGLEDEDEPSENSLEDEPSE 3545
Query: 229 NQ-KVEHESSLSPLEEKRFSK 248
N + E E S + LE++ K
Sbjct: 3546 NSLEDEDEPSENSLEDEPSEK 3608
Score = 28.1 bits (61), Expect = 5.7
Identities = 29/110 (26%), Positives = 52/110 (46%), Gaps = 4/110 (3%)
Frame = +3
Query: 40 AESSDDNDDEGPFFDLEFTAPEEE-EDGFEETNHKNQQQEEEKD-EDDDEDSDD--DDDD 95
+E +++DE LE E ED E + + + + EK ED+DE S+ +D+D
Sbjct: 3480 SEKGLEDEDEPSENSLEDEPSENSLEDEDEPSENSLEDEPSEKGLEDEDEPSEKGLEDED 3659
Query: 96 DSGETESEFKITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFILN 145
+ E E + PS N D + PSE+ ++ +L + + +L+
Sbjct: 3660 EPSEKGLEDED--EPSENSLEDED---EPSENDDYQNRLDSYQDRTGLLD 3794
Score = 27.7 bits (60), Expect = 7.5
Identities = 23/115 (20%), Positives = 50/115 (43%), Gaps = 1/115 (0%)
Frame = +3
Query: 46 NDDEGPFFDLEFTAPEE-EEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEF 104
++D P +EF + +E E G E+ + + E ED+ ++ +D+D+ E E
Sbjct: 3426 DEDMFPVDPIEFNSEDEPSEKGLEDED----EPSENSLEDEPSENSLEDEDEPSENSLED 3593
Query: 105 KITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPNNK 159
+ + +E + L ++ KG + + S L + + S + + N+
Sbjct: 3594 EPSEKGLEDEDEPSEKGLEDEDEPSEKGLEDEDEPSENSLEDEDEPSENDDYQNR 3758
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 41.2 bits (95), Expect = 7e-04
Identities = 28/110 (25%), Positives = 51/110 (45%), Gaps = 16/110 (14%)
Frame = +2
Query: 61 EEEEDGFEETNHKNQQQEEEKDEDDDE-------DSDDDDDDDSGETESE---------F 104
E+EED +E+ +++ E+ +++D+ E D +DDDDDD + E+E F
Sbjct: 614 EKEEDNYEKGEERDEADEDSEEDDELEKAGEFEMDDEDDDDDDEEDEEAEDMGNVRYEDF 793
Query: 105 KITLSPSNNERSDANLSLSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSS 154
+ ++R D L +S ED K + +S++ N+ S
Sbjct: 794 FGGKNEKGSKRKDQLLEVSGDEDDMESTKQKKRTASTYEKQRGKNSIKDS 943
Score = 38.5 bits (88), Expect = 0.004
Identities = 18/58 (31%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Frame = +2
Query: 47 DDEGPFFDLEFTAPEEEEDGFEETNHKNQQQ-EEEKDEDDDEDSDDDDDDDSGETESE 103
++E FD E +++ +G E+ K + E++ +E+DDED + +D+D E E E
Sbjct: 377 EEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDEDEEGSEDEDDEEDEKE 550
Score = 36.6 bits (83), Expect = 0.016
Identities = 17/55 (30%), Positives = 32/55 (57%)
Frame = +2
Query: 41 ESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDD 95
E SDD D+E D +F E+++ + ++E+++DE+ ED DD++D+
Sbjct: 380 EESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDEDEEGSEDEDDEEDE 544
Score = 33.9 bits (76), Expect = 0.10
Identities = 15/33 (45%), Positives = 21/33 (63%)
Frame = +2
Query: 73 KNQQQEEEKDEDDDEDSDDDDDDDSGETESEFK 105
K + EEE+ +D DE+ DDDDDD G + + K
Sbjct: 359 KRVELEEEESDDFDEELDDDDDDFEGVEKKKAK 457
Score = 29.3 bits (64), Expect = 2.6
Identities = 13/31 (41%), Positives = 19/31 (60%)
Frame = +2
Query: 69 ETNHKNQQQEEEKDEDDDEDSDDDDDDDSGE 99
E + + +EEE D+ D+E DDDDD + E
Sbjct: 350 EKKKRVELEEEESDDFDEELDDDDDDFEGVE 442
>TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 761
Score = 40.4 bits (93), Expect = 0.001
Identities = 19/60 (31%), Positives = 35/60 (57%)
Frame = +3
Query: 62 EEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESEFKITLSPSNNERSDANLS 121
++ G + N ++ EE++ED+DE+ +D DDD E+E EF S+ R+D+ ++
Sbjct: 339 KDTKGSDNDNDDEGKENEEEEEDEDEEEEDTSDDDGSESE-EFNADEEDSD*TRADSAIN 515
Score = 37.0 bits (84), Expect = 0.012
Identities = 22/52 (42%), Positives = 27/52 (51%)
Frame = +3
Query: 39 TAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSD 90
T S +DNDDEG E EE+ED EE + E E+ D+EDSD
Sbjct: 345 TKGSDNDNDDEGK----ENEEEEEDEDEEEEDTSDDDGSESEEFNADEEDSD 488
>TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-related
protein complex AP-3 beta 1 subunit {Mus musculus},
partial (2%)
Length = 931
Score = 40.0 bits (92), Expect = 0.001
Identities = 51/214 (23%), Positives = 81/214 (37%), Gaps = 7/214 (3%)
Frame = +1
Query: 39 TAESSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSG 98
TA SSD+ + E++ED E EEE DE+D S+D++++D+
Sbjct: 106 TASSSDEEQPPSTQKPADKVVNEQDEDSSSE--------EEEGDEEDGSSSEDEEEEDNQ 261
Query: 99 ETESEFKITLSPSNNERSDANLS-LSPSEDLFFKGKLVQLKSSSFILNNNNNNSSSSEPN 157
++ K + S N +S P+E S + + S SEP
Sbjct: 262 TQQTPSKDSKPASKNPPPSTPISNPKPAE-----------SESGSESGSESGTESDSEPE 408
Query: 158 NKPQFTASLLKSATKFRVFMSGLKKPKTDSVLEKKQHKPERIKFKVEEVPIVSLFTRDNS 217
+K + T K + S KP + Q P IK + V S N
Sbjct: 409 HKSEPTP---PPNPKVKPLAS---KPMKTQTQAQAQSTPVPIKSGTKRVAESS---TGND 561
Query: 218 SKNSNKSQT------NQNQKVEHESSLSPLEEKR 245
SK S K T + +VE ++ L+ + K+
Sbjct: 562 SKRSKKKTTTAGGGSDDENEVEEDAKLTGEDSKK 663
>TC77517 similar to PIR|T51159|T51159 HMG protein [imported] - Arabidopsis
thaliana, partial (70%)
Length = 907
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/51 (33%), Positives = 34/51 (66%)
Frame = +3
Query: 42 SSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDD 92
S+DD+++E D + + E++ E +H++ ++E+++DEDDDED + D
Sbjct: 495 SADDDEEES---DKDNSEVNNEDEASGEDDHQDDEEEDDEDEDDDEDDE*D 638
Score = 37.7 bits (86), Expect = 0.007
Identities = 18/58 (31%), Positives = 30/58 (51%)
Frame = +3
Query: 42 SSDDNDDEGPFFDLEFTAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGE 99
SS D+D+E E ++D E N E++ +D++ED +D+DDD+ E
Sbjct: 492 SSADDDEE-----------ESDKDNSEVNNEDEASGEDDHQDDEEEDDEDEDDDEDDE 632
Score = 37.4 bits (85), Expect = 0.009
Identities = 16/46 (34%), Positives = 28/46 (60%), Gaps = 2/46 (4%)
Frame = +3
Query: 58 TAPEEEEDGFEETNHKNQQQEE--EKDEDDDEDSDDDDDDDSGETE 101
+A ++EE+ ++ + N + E E D DDE+ DD+D+DD + E
Sbjct: 495 SADDDEEESDKDNSEVNNEDEASGEDDHQDDEEEDDEDEDDDEDDE 632
Score = 35.0 bits (79), Expect = 0.047
Identities = 11/46 (23%), Positives = 29/46 (62%)
Frame = +3
Query: 58 TAPEEEEDGFEETNHKNQQQEEEKDEDDDEDSDDDDDDDSGETESE 103
++ +++E+ ++ N + ++E EDD +D +++DD+D + E +
Sbjct: 492 SSADDDEEESDKDNSEVNNEDEASGEDDHQDDEEEDDEDEDDDEDD 629
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.304 0.123 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,644,230
Number of Sequences: 36976
Number of extensions: 132081
Number of successful extensions: 4177
Number of sequences better than 10.0: 348
Number of HSP's better than 10.0 without gapping: 1627
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2675
length of query: 374
length of database: 9,014,727
effective HSP length: 98
effective length of query: 276
effective length of database: 5,391,079
effective search space: 1487937804
effective search space used: 1487937804
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140913.4


