

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.3 - phase: 0
(476 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
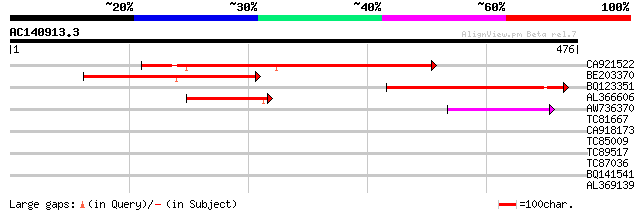
Score E
Sequences producing significant alignments: (bits) Value
CA921522 249 1e-66
BE203370 150 1e-36
BQ123351 108 6e-24
AL366606 106 2e-23
AW736370 49 5e-06
TC81667 similar to GP|8777488|dbj|BAA97068.1 contains similarity... 31 1.2
CA918173 weakly similar to GP|11034537|dbj putative glucosyl tra... 30 2.6
TC85009 similar to GP|14531294|gb|AAK66160.1 pectate lyase B {Fr... 29 3.5
TC89517 weakly similar to PIR|A85475|A85475 hypothetical protein... 29 3.5
TC87036 similar to GP|11138330|gb|AAG31326.1 putative serine/thr... 28 5.9
BQ141541 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, pa... 28 5.9
AL369139 weakly similar to GP|21554368|gb| unknown {Arabidopsis ... 28 7.7
>CA921522
Length = 767
Score = 249 bits (637), Expect = 1e-66
Identities = 128/256 (50%), Positives = 168/256 (65%), Gaps = 8/256 (3%)
Frame = +3
Query: 111 KTSDVSNHLITKSHIRGFTSKYLLDQANLSTTRQDTL------EAILALLIYGLLLFPNL 164
+T +V HLIT+ GF++ +L ++ TT D + +ILALL+YGL+LFP+L
Sbjct: 3 ETDEVKTHLITRGKFLGFSTDFLYER----TTFFDKMGVAYAFNSILALLVYGLVLFPSL 170
Query: 165 DNFVDMNAIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHLPSS-- 222
DNFVD+ AI+IF S+NPVPTLL DTY +IH RT GRG ILCC LLYRW SHLP +
Sbjct: 171 DNFVDIKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPR 350
Query: 223 FHDNSENWSYSQRIMALTPNEVVWLTPAAQVKEIIMGCGDFLNVPLLGIRGGINYNPELA 282
F N EN +S+R+M+LTP EVVW II CG F NVPLLG+ GGI+YNP LA
Sbjct: 351 FTTNPENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLA 530
Query: 283 MRQFGFPMKSKPINLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSVRHLGVRSGVAHEA 342
QFG+PM+ KP+++ ++ + TG R+ + AW +RR+ LG ++G +
Sbjct: 531 RHQFGYPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSS 710
Query: 343 YTQWVIDRAEEIGMPY 358
YTQWVIDRA +IGMPY
Sbjct: 711 YTQWVIDRAVQIGMPY 758
>BE203370
Length = 454
Score = 150 bits (378), Expect = 1e-36
Identities = 79/150 (52%), Positives = 100/150 (66%), Gaps = 2/150 (1%)
Frame = +1
Query: 63 CFTFPDFQLVPTLEAYSNLVGLPIAEKAPFTGPGTSLTPLVIAKDLYLKTSDVSNHLITK 122
CFTFPD+QL PTLE YS LVG P+ +K PFTG IA L+L+TS + L K
Sbjct: 4 CFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETSLIKAKLTIK 183
Query: 123 SHIRGFTSKYLLDQAN--LSTTRQDTLEAILALLIYGLLLFPNLDNFVDMNAIEIFHSKN 180
++ +K+L QA+ D +ILALLIYGL+LFPN+DN+VD++AI++F +KN
Sbjct: 184 GNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLTKN 363
Query: 181 PVPTLLADTYHAIHDRTLKGRGYILCCISL 210
PVPTLLAD YH+IHDR GRG IL C L
Sbjct: 364 PVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>BQ123351
Length = 725
Score = 108 bits (269), Expect = 6e-24
Identities = 60/153 (39%), Positives = 93/153 (60%)
Frame = +2
Query: 317 FMDAWSKVRRKSVRHLGVRSGVAHEAYTQWVIDRAEEIGMPYPAMRYVSSSTPSMPLPLL 376
F+ AW V ++ LG RS + +E+YT+WVIDRA + G+P+P R SS+T S LP+
Sbjct: 8 FIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLPMT 187
Query: 377 PATQDMYQEHLAMESREKQVWKARYNQAENLIMTLDGRDEQKTHENLMLKKELAKARREL 436
T++ Q LA +REK W+ RY +AE+ I TL G+ EQK HE L +++++ + L
Sbjct: 188 SKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDDLL 367
Query: 437 EEKDELLMRDSKRARGRRDFFARYCDSDSESDD 469
+EKD+LL + + + R D + DS+ +D
Sbjct: 368 QEKDKLLKKHITK-KQRMDSMDLFDGPDSDFED 463
>AL366606
Length = 393
Score = 106 bits (264), Expect = 2e-23
Identities = 56/78 (71%), Positives = 59/78 (74%), Gaps = 6/78 (7%)
Frame = -1
Query: 149 AILALLIYGLLLFPNLDNFVDMNAIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCI 208
AI L GL+LFPNLDNFVDMNAIE+FHSKNPVPTLLADTYHAIHDRTLKGRGYIL
Sbjct: 381 AIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYILWLH 202
Query: 209 SLL------YRWFISHLP 220
L +R F S LP
Sbjct: 201 ILFSNRVVYFRTFRSSLP 148
>AW736370
Length = 488
Score = 48.5 bits (114), Expect = 5e-06
Identities = 29/90 (32%), Positives = 43/90 (47%)
Frame = -2
Query: 368 TPSMPLPLLPATQDMYQEHLAMESREKQVWKARYNQAENLIMTLDGRDEQKTHENLMLKK 427
TP+ PLP+ T+ YQE L RE WK Y + + T+ G EQ+ +++
Sbjct: 481 TPAPPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*KDV 302
Query: 428 ELAKARRELEEKDELLMRDSKRARGRRDFF 457
+AK ++E D L R R + R D F
Sbjct: 301 IIAKLNERIKENDAALDRIPGRKKRRMDLF 212
>TC81667 similar to GP|8777488|dbj|BAA97068.1 contains similarity to SF16
protein~gene_id:K15M2.19 {Arabidopsis thaliana}, partial
(34%)
Length = 1216
Score = 30.8 bits (68), Expect = 1.2
Identities = 18/59 (30%), Positives = 25/59 (41%)
Frame = -2
Query: 273 GGINYNPELAMRQFGFPMKSKPINLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSVRH 331
GG+N NP FP L +P F+T A K + W + R+S+RH
Sbjct: 780 GGLNLNPN-------FPRTRCNPTLHPAPTPAFFTKAIRSLTKILVTVWPPLMRESIRH 625
>CA918173 weakly similar to GP|11034537|dbj putative glucosyl transferase
{Oryza sativa (japonica cultivar-group)}, partial (19%)
Length = 769
Score = 29.6 bits (65), Expect = 2.6
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = -3
Query: 49 LVHTLVQFYDPSFRCFTFPDFQ 70
L+HTLVQ + PS RCF FP+++
Sbjct: 113 LIHTLVQKW-PS*RCFPFPNYE 51
>TC85009 similar to GP|14531294|gb|AAK66160.1 pectate lyase B {Fragaria x
ananassa}, partial (55%)
Length = 914
Score = 29.3 bits (64), Expect = 3.5
Identities = 12/51 (23%), Positives = 26/51 (50%)
Frame = +3
Query: 304 FFYTNAPTGQRKAFMDAWSKVRRKSVRHLGVRSGVAHEAYTQWVIDRAEEI 354
F +TNA ++ K++ +S+ + + ++H+A + +D EEI
Sbjct: 138 FAFTNAIDSNESRVVNGEEKLKMQSLNNSSMAESLSHDAINEHAVDNPEEI 290
>TC89517 weakly similar to PIR|A85475|A85475 hypothetical protein AT4g40080
[imported] - Arabidopsis thaliana, partial (46%)
Length = 1177
Score = 29.3 bits (64), Expect = 3.5
Identities = 23/87 (26%), Positives = 34/87 (38%), Gaps = 1/87 (1%)
Frame = -3
Query: 161 FPNLDNFVDMNAIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHLP 220
F L F++ N H NP+ +H IH Y+ + L W I P
Sbjct: 764 FLPLPKFINSNQ-NFIHHHNPI------IFHQIH--------YLNNHLVLFTHWSIRFFP 630
Query: 221 SSFHDNSENWSYS-QRIMALTPNEVVW 246
+SFH + S+S Q + N +W
Sbjct: 629 NSFHQGQQRISFS*QITIGKIRNSFLW 549
>TC87036 similar to GP|11138330|gb|AAG31326.1 putative serine/threonine
kinase GDBrPK {Vitis vinifera}, partial (66%)
Length = 1212
Score = 28.5 bits (62), Expect = 5.9
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = +2
Query: 214 WFISHLPSSFHDNSENWS 231
WF+ +LP F ++ ENWS
Sbjct: 692 WFLKNLPLEFMEDGENWS 745
>BQ141541 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, partial (2%)
Length = 1320
Score = 28.5 bits (62), Expect = 5.9
Identities = 18/62 (29%), Positives = 32/62 (51%)
Frame = +1
Query: 414 RDEQKTHENLMLKKELAKARRELEEKDELLMRDSKRARGRRDFFARYCDSDSESDDPPTT 473
R E + +E+ + ++E RRE E++ R+++ ARG R R + DS + D T
Sbjct: 751 RTEDRKYEHRIAERE---RRREGEKEASNTRREARSARGGRRGNVRRAEEDSGTKDTATA 921
Query: 474 SY 475
+
Sbjct: 922 RH 927
>AL369139 weakly similar to GP|21554368|gb| unknown {Arabidopsis thaliana},
partial (11%)
Length = 388
Score = 28.1 bits (61), Expect = 7.7
Identities = 22/62 (35%), Positives = 27/62 (43%), Gaps = 7/62 (11%)
Frame = +3
Query: 414 RDEQKTHENLMLKKEL-------AKARRELEEKDELLMRDSKRARGRRDFFARYCDSDSE 466
RD +KT EN + E K RE E D D +R R R+ +SDSE
Sbjct: 66 RDSRKTRENSDIDTETDRKHISSKKKSRESSESDS----DKRRRRKRKSRRKYSSESDSE 233
Query: 467 SD 468
SD
Sbjct: 234 SD 239
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,824,265
Number of Sequences: 36976
Number of extensions: 207305
Number of successful extensions: 1177
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1170
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1174
length of query: 476
length of database: 9,014,727
effective HSP length: 100
effective length of query: 376
effective length of database: 5,317,127
effective search space: 1999239752
effective search space used: 1999239752
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140913.3


