

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140849.12 + phase: 0
(256 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
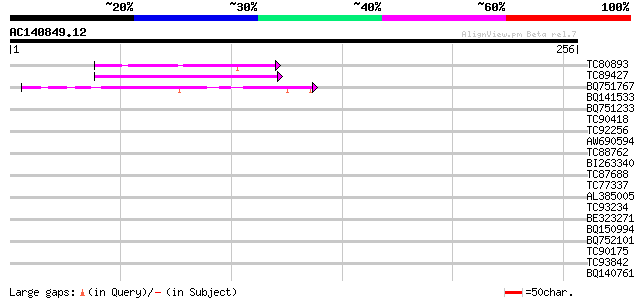
Score E
Sequences producing significant alignments: (bits) Value
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 46 1e-05
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 43 1e-04
BQ751767 similar to SP|O10302|GTA_N Probable global transactivat... 40 7e-04
BQ141533 similar to GP|15278108|gb cystathionine beta-lyase {Neu... 37 0.006
BQ751233 similar to GP|18307448|em probable low-affinity hexose ... 37 0.006
TC90418 similar to GP|11762174|gb|AAG40365.1 At1g17620 {Arabidop... 37 0.007
TC92256 similar to PIR|T49842|T49842 related to Na+/H+-exchangin... 36 0.013
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 36 0.017
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 36 0.017
BI263340 weakly similar to PIR|T49223|T492 uclacyanin 3 [importe... 35 0.028
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 35 0.028
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 35 0.037
AL385005 weakly similar to GP|21438547|emb Ratten cDNA {Rattus n... 35 0.037
TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 34 0.048
BE323271 similar to PIR|T52305|T523 En/Spm-like transposon prote... 34 0.048
BQ150994 similar to GP|22122173|dbj preproneuropeptide B {Homo s... 34 0.063
BQ752101 similar to PIR|S57177|S57 branched-chain-amino-acid tra... 34 0.063
TC90175 weakly similar to GP|6002488|gb|AAF00009.1| hypothetical... 34 0.063
TC93842 weakly similar to GP|13365999|dbj|BAB39276. hypothetical... 33 0.11
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 33 0.11
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 46.2 bits (108), Expect = 1e-05
Identities = 35/85 (41%), Positives = 51/85 (59%), Gaps = 1/85 (1%)
Frame = -1
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
GWG SSSS+S++ SS S +++ +SSPS S+ PSS P SGS S ++ S
Sbjct: 571 GWGVSSSSSSSSS--SSSSSPSSSSSSSPSSSSSSPSS--LPPAPLASGSSSGSPSSSPS 404
Query: 99 AWG-SSSRPSSASGPPTSNQTSQTS 122
+ SSS PSS+S +S+ +S +S
Sbjct: 403 SPE*SSSSPSSSSSCTSSSSSSSSS 329
Score = 35.4 bits (80), Expect = 0.022
Identities = 27/92 (29%), Positives = 45/92 (48%), Gaps = 2/92 (2%)
Frame = -1
Query: 49 SNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSS 108
+ WG S S ++++ +SS S PSS S + S+S S + ++ SS PSS
Sbjct: 577 AGGWGVSSSSSSSSSSSSSS-----PSSSSSSSPSSSSSSPSSLPPAPLASGSSSGSPSS 413
Query: 109 ASGPP--TSNQTSQTSLRPRSAETRPGSSHLS 138
+ P +S+ S +S S+ + SS +S
Sbjct: 412 SPSSPE*SSSSPSSSSSCTSSSSSSSSSSSVS 317
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 42.7 bits (99), Expect = 1e-04
Identities = 29/85 (34%), Positives = 48/85 (56%)
Frame = -2
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
G S SSSS+S++ S S ++++ +SS S S P+ G+G+ ++ S +S ++S
Sbjct: 545 G*SSSSSSSSSSSSSSPSSLSSSSSSSSSSSSSLPPAPFGTGSSSGSASSSSSSPEKSSS 366
Query: 99 AWGSSSRPSSASGPPTSNQTSQTSL 123
SSS SS+S +S+ S SL
Sbjct: 365 LSSSSSFTSSSSSSSSSSSVSLPSL 291
Score = 31.2 bits (69), Expect = 0.41
Identities = 24/73 (32%), Positives = 36/73 (48%), Gaps = 1/73 (1%)
Frame = -2
Query: 77 GGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRP-GSS 135
GG + S+S S +S ++ S+ SSS SS+S PP T +S S+ + P SS
Sbjct: 548 GG*SSSSSSSSSSSSSSPSSLSSSSSSSSSSSSSLPPAPFGTGSSSGSASSSSSSPEKSS 369
Query: 136 HLSRFAEHVGENS 148
LS + +S
Sbjct: 368 SLSSSSSFTSSSS 330
>BQ751767 similar to SP|O10302|GTA_N Probable global transactivator. [OpMNPV]
{Orgyia pseudotsugata multicapsid polyhedrosis virus},
partial (7%)
Length = 771
Score = 40.4 bits (93), Expect = 7e-04
Identities = 46/142 (32%), Positives = 62/142 (43%), Gaps = 8/142 (5%)
Frame = +2
Query: 6 RRWTSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGAS 65
RRWT S RR T + P + PS G +++S+ ++ SS ++A
Sbjct: 383 RRWTRSRRR---TWW*RRTTP---SRPSSTGR----NRASTRSAGTESSSTRRTSSAARP 532
Query: 66 SPSHLSARPS----SGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQT 121
PS A PS G S RPS +G T +W PSSA P T+ +S
Sbjct: 533 RPSTRPAAPSRPTRDGASPARPSRTGCP-----TLAPSW-----PSSAPAPLTTRASSAA 682
Query: 122 SLR---PRSAETRPGS-SHLSR 139
+ R R+A TR GS H SR
Sbjct: 683 TSRSPLSRAARTRTGSREHSSR 748
>BQ141533 similar to GP|15278108|gb cystathionine beta-lyase {Neurospora
crassa}, partial (44%)
Length = 609
Score = 37.4 bits (85), Expect = 0.006
Identities = 34/126 (26%), Positives = 49/126 (37%)
Frame = +2
Query: 10 SSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSH 69
S+TR P ++L S G S+ S+SA W SS P+
Sbjct: 176 STTRHSATGFTSPSTHPAAVSLHSTHGCFSAASRRSASA---WKSS----------RPTP 316
Query: 70 LSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAE 129
++ PSS + +R +T T T AW + P S TS S +S PRS
Sbjct: 317 CASPPSSSRTVSRCATPA*SRIRSTTCTGAWRGARAPCCPSRLATSPYPSASSRAPRSGP 496
Query: 130 TRPGSS 135
+ S+
Sbjct: 497 SASPSA 514
>BQ751233 similar to GP|18307448|em probable low-affinity hexose transporter
HXT3 {Neurospora crassa}, partial (30%)
Length = 659
Score = 37.4 bits (85), Expect = 0.006
Identities = 39/116 (33%), Positives = 52/116 (44%), Gaps = 3/116 (2%)
Frame = +1
Query: 14 RGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSS---SASNAWGSSLSPNANAGASSPSHL 70
R +V AV +LP TL + S S + SS S + G SSPS +
Sbjct: 304 RAPSSVCSASAVSSVPSLPVSSPTLSAAA*PSPPLPSGPASVSSSRSRPSPPGTSSPSAV 483
Query: 71 SARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPR 126
S+ S+ S PS+S S L +SA SSSRP+S+S P S + PR
Sbjct: 484 SSTVSA--SAPSPSSSPCTSPSPLRPSSA-ASSSRPTSSSSPSASGPPRWVTGAPR 642
>TC90418 similar to GP|11762174|gb|AAG40365.1 At1g17620 {Arabidopsis
thaliana}, partial (9%)
Length = 835
Score = 37.0 bits (84), Expect = 0.007
Identities = 40/119 (33%), Positives = 50/119 (41%), Gaps = 1/119 (0%)
Frame = +1
Query: 24 AVPKPINL-PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTR 82
AVP + PS G + S SSSS++ S S SPSHLS P+S
Sbjct: 172 AVPAAVAAAPSSSGFSSYFSSSSSSSA*PAPLSTSSTVPTVLPSPSHLSNFPTSTSPPPP 351
Query: 83 PSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFA 141
PST S S T ++ PS + PP SQ SL ++ P HLS A
Sbjct: 352 PSTQSSTSTSLQKTL----TNISPSFTNQPP-----SQFSLTTSTSVKEP--YHLSNMA 495
>TC92256 similar to PIR|T49842|T49842 related to Na+/H+-exchanging protein
[imported] - Neurospora crassa, partial (43%)
Length = 1166
Score = 36.2 bits (82), Expect = 0.013
Identities = 36/140 (25%), Positives = 57/140 (40%), Gaps = 3/140 (2%)
Frame = +1
Query: 26 PKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPST 85
P+ ++ S+ + W S SSSS+ SS A + SP S+ PS G RP+
Sbjct: 301 PRKTSIASRSHSRAWCSASSSSSP---ASSCPAGTYAPSGSPWPYSSAPS*PACGWRPAC 471
Query: 86 SGSDMASELTTTSAWGSSS--RPSSASGPP-TSNQTSQTSLRPRSAETRPGSSHLSRFAE 142
+ + W S+ RP + P S +S T++ RS T +S
Sbjct: 472 LFTPWCRTFRSCMLWPSAPA*RPPTRCCPT**SRASSPTTISQRSCRTSSSASRAPMTDS 651
Query: 143 HVGENSVAWNGARTTETLGI 162
+ +S+ + T T GI
Sbjct: 652 GIRSSSLRCT*SSTLATAGI 711
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 35.8 bits (81), Expect = 0.017
Identities = 24/65 (36%), Positives = 37/65 (56%)
Frame = -3
Query: 71 SARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
S+ PSS S + S+S S +S + +S+ SSS SS+S +S+ +S +S S+ T
Sbjct: 247 SSSPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSSCSSSSSSSCSS---SSSST 77
Query: 131 RPGSS 135
P SS
Sbjct: 76 SPSSS 62
Score = 35.0 bits (79), Expect = 0.028
Identities = 27/76 (35%), Positives = 40/76 (52%)
Frame = -3
Query: 47 SASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRP 106
S N GS + +SSPS S+ SS S + S+S S +S +++S+ SSS
Sbjct: 298 SICNNSGSGRVLILSISSSSPSSSSS-SSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSSC 122
Query: 107 SSASGPPTSNQTSQTS 122
SS+S S+ +S TS
Sbjct: 121 SSSSSSSCSSSSSSTS 74
Score = 27.3 bits (59), Expect = 5.9
Identities = 18/55 (32%), Positives = 29/55 (52%)
Frame = -3
Query: 84 STSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
+ SGS L+ +S+ SSS SS+S +S+ +S +S S+ + SS S
Sbjct: 289 NNSGSGRVLILSISSSSPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSS 125
Score = 26.9 bits (58), Expect = 7.7
Identities = 15/38 (39%), Positives = 23/38 (60%)
Frame = -3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGS 79
S SSSS+S++ SS ++++ + S S S PSS S
Sbjct: 169 SSSSSSSSSSSSSSSCSSSSSSSCSSSSSSTSPSSSSS 56
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 35.8 bits (81), Expect = 0.017
Identities = 32/116 (27%), Positives = 54/116 (45%), Gaps = 6/116 (5%)
Frame = +3
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARP--SSGGSGTRPSTSG----SDMASELTTT 97
SS SAS+ ++ SP A+ ASSP + P S+ S P+T+ + +S ++T
Sbjct: 99 SSPSASSPVSTTTSPPASTPASSPVSTTTSPPASTPASSPVPTTTSPPAPTPASSPVSTN 278
Query: 98 SAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNG 153
S S + A+ P+ + T+ S P S SS+ + +G S + +G
Sbjct: 279 SPTASPAGSLPAAATPSPSSTTAGSPSPSSTPPGTRSSNETTPGRSIGVASASPSG 446
>BI263340 weakly similar to PIR|T49223|T492 uclacyanin 3 [imported] -
Arabidopsis thaliana, partial (14%)
Length = 681
Score = 35.0 bits (79), Expect = 0.028
Identities = 30/89 (33%), Positives = 38/89 (41%), Gaps = 8/89 (8%)
Frame = +3
Query: 55 SLSPNANAGASSPSHLSARPSSGGSG----TRPSTSGSDMASELTTTSAWGSSSRPSSAS 110
S SP+ + +S PSH S+ P S G G T PST +TS + PS+ S
Sbjct: 168 STSPS-HGSSSPPSHGSSSPPSHGGGYYTPTPPSTGCGYSPPHDPSTSTPSHNQTPSTPS 344
Query: 111 GPPTS----NQTSQTSLRPRSAETRPGSS 135
PP+S N T P T P S
Sbjct: 345 NPPSSGGYYNSPPSTPTDPPVTLTPPSPS 431
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 35.0 bits (79), Expect = 0.028
Identities = 27/84 (32%), Positives = 45/84 (53%), Gaps = 5/84 (5%)
Frame = -2
Query: 30 NLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS-----HLSARPSSGGSGTRPS 84
NL S L + SSSS+S++ SS S ++++ +SSPS + PS S + S
Sbjct: 582 NLSSIPPPLTFSFSSSSSSSSSLPSSSSSSSSSKSSSPSLPPFAFFFSTPSKSSSSSSNS 403
Query: 85 TSGSDMASELTTTSAWGSSSRPSS 108
+S S ++S +T + S+ P+S
Sbjct: 402 SSKSSLSSSSNSTRFFFSNFFPTS 331
Score = 31.6 bits (70), Expect = 0.31
Identities = 34/101 (33%), Positives = 53/101 (51%), Gaps = 17/101 (16%)
Frame = -2
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSS--------GGSGTRPSTSGS----- 88
S SSSS+S++ SS+S + +SS S LS+ SS S ++ S + S
Sbjct: 756 SSSSSSSSSSSSSSISNSPAFSSSSSSSLSSSASSLSSPFS*LSSSFSKYSVNSSIFKNL 577
Query: 89 -DMASELT---TTSAWGSSSRPSSASGPPTSNQTSQTSLRP 125
+ LT ++S+ SSS PSS+S +S+++S SL P
Sbjct: 576 SSIPPPLTFSFSSSSSSSSSLPSSSSS-SSSSKSSSPSLPP 457
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 34.7 bits (78), Expect = 0.037
Identities = 30/75 (40%), Positives = 42/75 (56%)
Frame = -2
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
G K+SSS+S++ SSLS +S+PS S+ P S S S+S S +S ++ S+
Sbjct: 757 GCKASSSSSSS-SSSLS-----SSSTPSSSSSSPPSSSSS---SSSSSSSSSSWSSPSSS 605
Query: 101 GSSSRPSSASGPPTS 115
SSS SSA P S
Sbjct: 604 SSSSPFSSAPPAPPS 560
Score = 33.5 bits (75), Expect = 0.082
Identities = 19/59 (32%), Positives = 35/59 (59%)
Frame = -2
Query: 52 WGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSAS 110
+G + ++++ +SS S+ PSS S + P +S S +S +++S+W S S SS+S
Sbjct: 763 FGGCKASSSSSSSSSSLSSSSTPSS--SSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSS 593
Score = 32.7 bits (73), Expect = 0.14
Identities = 23/60 (38%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Frame = -2
Query: 80 GTRPSTSGSDMASELTTTSAWGSSSRPSS-ASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
G + S+S S +S L+ SSS PSS +S PP+S+ +S +S S+ + P SS S
Sbjct: 757 GCKASSSSSSSSSSLS------SSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSS 596
Score = 30.8 bits (68), Expect = 0.53
Identities = 29/93 (31%), Positives = 43/93 (46%), Gaps = 12/93 (12%)
Frame = -2
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGT---------- 81
PS + S SSSS+S++ SS S +++ +SSP S+ P + SG
Sbjct: 697 PSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSP--FSSAPPAPPSGAFFELLLLLEL 524
Query: 82 --RPSTSGSDMASELTTTSAWGSSSRPSSASGP 112
P S D +S + + SSS SS+S P
Sbjct: 523 LPYPPPSSDDNSSSPSPNAPSPSSSSSSSSSLP 425
Score = 30.0 bits (66), Expect = 0.91
Identities = 17/54 (31%), Positives = 29/54 (53%)
Frame = -2
Query: 77 GGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
GG S+S S + ++T + SSS PSS+S +S+ +S + P S+ +
Sbjct: 760 GGCKASSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSS 599
Score = 27.7 bits (60), Expect = 4.5
Identities = 18/69 (26%), Positives = 34/69 (49%)
Frame = -2
Query: 60 ANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTS 119
+++ +SS S LS+ + S + P +S S +S +++S+W S +S +S +S
Sbjct: 745 SSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWS-----SPSSSSSSSPFSS 581
Query: 120 QTSLRPRSA 128
P A
Sbjct: 580 APPAPPSGA 554
>AL385005 weakly similar to GP|21438547|emb Ratten cDNA {Rattus norvegicus},
partial (5%)
Length = 469
Score = 34.7 bits (78), Expect = 0.037
Identities = 24/68 (35%), Positives = 29/68 (42%), Gaps = 4/68 (5%)
Frame = +2
Query: 53 GSSLSPNANAG----ASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSS 108
G LS N +G A+SPS ++ PS T PS SGS T SS P
Sbjct: 20 GQKLSINVGSGSNSPATSPSPYASSPSPSTGATPPSASGSPSPGSPVTP----SSQSPGG 187
Query: 109 ASGPPTSN 116
+ PP N
Sbjct: 188 SVSPPPEN 211
>TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (11%)
Length = 591
Score = 34.3 bits (77), Expect = 0.048
Identities = 32/124 (25%), Positives = 50/124 (39%), Gaps = 11/124 (8%)
Frame = +2
Query: 22 KVAVPK----PINLPSQRGTLGWGSKSSSSASNAWG-----SSLSPNANAGASSPSHLSA 72
KVA+P P +P + KSS ++ W S + A+A +S S + +
Sbjct: 20 KVAIPSDQSIPQFVPPNKNCHPKSPKSSRNSQQKWSRTS*SSERATPASASLTSSSSIPS 199
Query: 73 RPS--SGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
+ S S +RP + + A +S S + PSS P S T +T SA
Sbjct: 200 QRSRTSRSPSSRPRHTSTGTAPPFAVSSPASSPTTPSSTRSSPGSTSTPRTPSTSFSARP 379
Query: 131 RPGS 134
P +
Sbjct: 380 PPST 391
Score = 32.7 bits (73), Expect = 0.14
Identities = 31/130 (23%), Positives = 56/130 (42%)
Frame = +2
Query: 6 RRWTSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGAS 65
++W+ ++ ++ ++PSQR S SS + G++ ++ AS
Sbjct: 116 QKWSRTS*SSERATPASASLTSSSSIPSQRSRTS-RSPSSRPRHTSTGTAPPFAVSSPAS 292
Query: 66 SPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRP 125
SP+ S+ SS GS + P T + S+RP ++ PPT ++ T +
Sbjct: 293 SPTTPSSTRSSPGSTSTPRTPSTSF------------SARPPPSTPPPTPFRSRPT--KG 430
Query: 126 RSAETRPGSS 135
S + P SS
Sbjct: 431 SSPSSTPSSS 460
>BE323271 similar to PIR|T52305|T523 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 431
Score = 34.3 bits (77), Expect = 0.048
Identities = 29/86 (33%), Positives = 38/86 (43%), Gaps = 4/86 (4%)
Frame = +3
Query: 55 SLSPNANAGASSPSHLSARPSSGGSG----TRPSTSGSDMASELTTTSAWGSSSRPSSAS 110
S SP+ + +S PSH S+ P S G G T PST +TS + PS+ S
Sbjct: 159 STSPS-HGSSSPPSHGSSSPPSHGGGYYTPTPPSTGCGYSPPHDPSTSTPSHNQTPSTPS 335
Query: 111 GPPTSNQTSQTSLRPRSAETRPGSSH 136
PP+S + P S T P H
Sbjct: 336 NPPSSGGYYNS---PPSTPTDPPE*H 404
>BQ150994 similar to GP|22122173|dbj preproneuropeptide B {Homo sapiens},
partial (15%)
Length = 1036
Score = 33.9 bits (76), Expect = 0.063
Identities = 26/99 (26%), Positives = 38/99 (38%)
Frame = +2
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
PS+RGT G++ S+ +PNA + P H P SD A
Sbjct: 329 PSRRGTRTRGNEKRSTPKTRPAEKRNPNAGRAPAPPGH------------NPPRETSDEA 472
Query: 92 SELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
T +++R + + PP + S PRS ET
Sbjct: 473 RR-TQARREKTTNRARTKTKPPRKKTRQERSSAPRSGET 586
>BQ752101 similar to PIR|S57177|S57 branched-chain-amino-acid transaminase
(EC 2.6.1.42) BAT2 cytosolic - yeast, partial (46%)
Length = 720
Score = 33.9 bits (76), Expect = 0.063
Identities = 26/87 (29%), Positives = 42/87 (47%)
Frame = -1
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S S+ A + SS P+A + +RP + S P+ S + +++ TT A
Sbjct: 696 SASALPAPPCFTSSPPPSAPTTLPASKPSPSRPPTTPSALGPAVSATKSSAQ-TTPPASC 520
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSA 128
SSRP +A+ T +++ S PRSA
Sbjct: 519 RSSRPPAAATSRTCGSSARRSTSPRSA 439
>TC90175 weakly similar to GP|6002488|gb|AAF00009.1| hypothetical protein
{Penicillium chrysogenum}, partial (87%)
Length = 1290
Score = 33.9 bits (76), Expect = 0.063
Identities = 28/89 (31%), Positives = 43/89 (47%), Gaps = 11/89 (12%)
Frame = -3
Query: 55 SLSPNANAGASSPSHLSARPS-----SGGSGTR----PSTSGSDMASELTTTSAWGSSSR 105
S SP+ + ASSP+ +ARPS S S TR PS + A ++ +A ++
Sbjct: 901 SASPSRASPASSPTAPTARPSTSTTTSANSSTRRPGRPSPRPASPACPSSSAAAPSPRAK 722
Query: 106 PSSASG--PPTSNQTSQTSLRPRSAETRP 132
PS ++G PP + T + P +A P
Sbjct: 721 PSGSAGRRPPRAGTTPSSCRPPTTAACCP 635
Score = 33.5 bits (75), Expect = 0.082
Identities = 28/94 (29%), Positives = 37/94 (38%)
Frame = -3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S+ S S++ S+ ++SPS S S RPSTS TTSA
Sbjct: 967 SRQSPSSAQTTRSTRRRRPPTPSASPSRASPASSPTAPTARPSTST--------TTSANS 812
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
S+ RP S P S +S S +P S
Sbjct: 811 STRRPGRPSPRPASPACPSSSAAAPSPRAKPSGS 710
Score = 27.3 bits (59), Expect = 5.9
Identities = 17/59 (28%), Positives = 28/59 (46%)
Frame = -3
Query: 75 SSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPG 133
+S S + T+ S T SA S + P+S+ PT+ ++ T+ S+ RPG
Sbjct: 970 TSRQSPSSAQTTRSTRRRRPPTPSASPSRASPASSPTAPTARPSTSTTTSANSSTRRPG 794
>TC93842 weakly similar to GP|13365999|dbj|BAB39276. hypothetical protein
{Oryza sativa (japonica cultivar-group)}, partial (12%)
Length = 697
Score = 33.1 bits (74), Expect = 0.11
Identities = 29/107 (27%), Positives = 48/107 (44%)
Frame = -3
Query: 29 INLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGS 88
++ R T S ++ ++++ +S S S+P+ LS + PS+S
Sbjct: 671 VSATRSRATRTLPSSATCTSASCPRTSTSATTPRRTSTPASLSTASCTPRWRRGPSSSRK 492
Query: 89 DMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
A +TSA S RP+ SGPP +S++ RP A P S+
Sbjct: 491 RCARSSRSTSA--RSRRPAPRSGPP----SSRSRKRPPRAGATPASA 369
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus
norvegicus}, partial (9%)
Length = 620
Score = 33.1 bits (74), Expect = 0.11
Identities = 30/108 (27%), Positives = 50/108 (45%), Gaps = 1/108 (0%)
Frame = +2
Query: 26 PKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPS-SGGSGTRPS 84
P P R + +SSS S++ ++ P A + AS P S PS + TR +
Sbjct: 68 PSPTRRAPPRPKTSRPTSTSSSPSSSRRATTRPRAPSQASPPGPPSRPPSPMPWT*TRRA 247
Query: 85 TSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRP 132
+ + + + T+A +++ + A+ T QT T+LR RS RP
Sbjct: 248 ATAATRTAAIAVTAATRTATPATIATTAATRAQT--TALRTRSLYLRP 385
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.125 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,742,420
Number of Sequences: 36976
Number of extensions: 134841
Number of successful extensions: 1363
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 1199
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1302
length of query: 256
length of database: 9,014,727
effective HSP length: 94
effective length of query: 162
effective length of database: 5,538,983
effective search space: 897315246
effective search space used: 897315246
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC140849.12


