

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140774.1 - phase: 0
(505 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
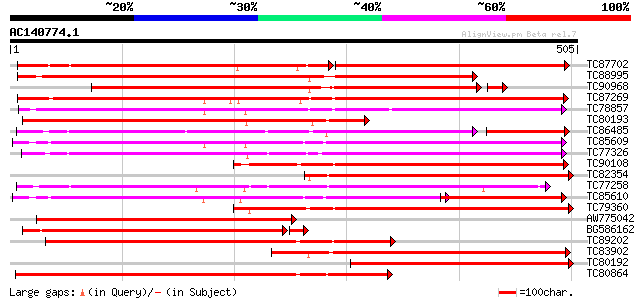
Score E
Sequences producing significant alignments: (bits) Value
TC87702 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 7... 275 e-142
TC88995 similar to SP|O81971|C7D9_SOYBN Cytochrome P450 71D9 (EC... 474 e-134
TC90968 similar to SP|O81971|C7D9_SOYBN Cytochrome P450 71D9 (EC... 384 e-112
TC87269 similar to PIR|T05940|T05940 cytochrome P450 83D1p - soy... 397 e-111
TC78857 weakly similar to SP|O81974|C7D8_SOYBN Cytochrome P450 7... 346 1e-95
TC80193 weakly similar to SP|O22307|C7DB_LOTJA Cytochrome P450 7... 338 4e-93
TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from p... 270 7e-93
TC85609 weakly similar to SP|Q9LTL2|C72P_ARATH Cytochrome P450 7... 336 1e-92
TC77326 similar to GP|12004682|gb|AAG44132.1 cytochrome P450 {Pi... 335 2e-92
TC90108 similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (E... 332 2e-91
TC82354 similar to SP|O22307|C7DB_LOTJA Cytochrome P450 71D11 (E... 328 2e-90
TC77258 similar to GP|5059126|gb|AAD38930.1| cytochrome P450 mon... 326 1e-89
TC85610 similar to GP|21068674|emb|CAD31843. putative cytochrome... 223 6e-89
TC79360 similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (E... 323 7e-89
AW775042 weakly similar to SP|O22307|C7DB Cytochrome P450 71D11 ... 312 2e-85
BG586162 weakly similar to SP|O22307|C7DB Cytochrome P450 71D11 ... 307 8e-85
TC89202 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 7... 305 3e-83
TC83902 similar to SP|O81974|C7D8_SOYBN Cytochrome P450 71D8 (EC... 301 3e-82
TC80192 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 7... 301 5e-82
TC80864 similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (E... 300 7e-82
>TC87702 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (EC
1.14.-.-). [Soybean] {Glycine max}, partial (51%)
Length = 1732
Score = 275 bits (702), Expect(2) = e-142
Identities = 128/209 (61%), Positives = 168/209 (80%), Gaps = 1/209 (0%)
Frame = +2
Query: 291 TDNNIKAIILDIIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYI 350
++N +KA I DI GAG +T+ TT+ WAM+EL+++P+VMKKAQ EVR ++N+KG V E +
Sbjct: 896 SNNVLKAKISDIFGAGSDTTFTTLEWAMSELIKNPQVMKKAQAEVRSVYNEKGYVDEASL 1075
Query: 351 NELEYLKLVVKETLRLHPPTPLLL-RECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWT 409
++L+YLK V+ ETLRLH P PLLL R+C + CEI GY IPAKSKVIVNAW+I RD +YW
Sbjct: 1076 HKLKYLKSVITETLRLHAPIPLLLPRQCSEKCEINGYEIPAKSKVIVNAWSICRDSRYWI 1255
Query: 410 EPERFHPERFIGSSIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRLP 469
E E+F PERFI SS+DYKG +F++IPFGAGRR+CPG+T G+ ++E++LA LL+HFDWR+P
Sbjct: 1256 EAEKFFPERFIDSSVDYKGVDFQFIPFGAGRRMCPGMTSGIASLEISLANLLFHFDWRMP 1435
Query: 470 NGMKGEDLDMTEQFGANVKRKSDLYLIPT 498
NG +DLDM E FG V+RK DL L+PT
Sbjct: 1436 NGNNADDLDMDESFGLAVRRKHDLRLVPT 1522
Score = 249 bits (635), Expect(2) = e-142
Identities = 133/286 (46%), Positives = 195/286 (67%), Gaps = 5/286 (1%)
Frame = +1
Query: 8 LLALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAK 67
L++ +L F+ +V+ R+ KK++ LPPGP KLP+IG+IH L + PHQ L LA+
Sbjct: 46 LISTILGFLLFMVIKFIWRSKTKKTTY-KLPPGPRKLPLIGNIHQL-GTLPHQALAKLAQ 219
Query: 68 VYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYG 127
YG LMH+QLGE+S IVVSS E A+E+MKTHD+ FA++P +++ +I+ YG + FSP+G
Sbjct: 220 EYGSLMHMQLGELSCIVVSSQEMAKEIMKTHDLNFANRPPLLSAEIVTYGYKGMTFSPHG 399
Query: 128 NYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTIT 187
+YWRQ+RKICT+ELL+Q RV SFR REEE N +K I+S +GSPIN+++ + + +T
Sbjct: 400 SYWRQMRKICTMELLSQNRVESFRLQREEELANFVKDINSSEGSPINLSEKLDSLAYGLT 579
Query: 188 TRAAFGNKCKGQEQ---LVSLANGESVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQI 244
+R AFG + +++ + + V GGF + +L+PS + LQ+++G R +IE LH +
Sbjct: 580 SRTAFGANVEDEDKEKYRKIIKDVLKVAGGFSLADLYPSIRILQVLTGLRQRIEKLHGET 759
Query: 245 DLILVNIINEH--KEAKSKAKEGEVEEDLVDVLQKFQGGNDIDQDI 288
D IL NI+ H K ++KAKE E EDLVDVL K Q +D++ I
Sbjct: 760 DKILENIVRSHRKKNLETKAKE-EKWEDLVDVLLKLQTNSDLEHPI 894
>TC88995 similar to SP|O81971|C7D9_SOYBN Cytochrome P450 71D9 (EC 1.14.-.-)
(P450 CP3). [Soybean] {Glycine max}, partial (71%)
Length = 1253
Score = 474 bits (1219), Expect = e-134
Identities = 239/415 (57%), Positives = 309/415 (73%), Gaps = 6/415 (1%)
Frame = +1
Query: 8 LLALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAK 67
+ +L++F +LTK K +SSA NLPPGP KLPIIG+IH+L+ S PH +LRDL+
Sbjct: 31 IFSLLMFVFIANKILTK----KSESSAQNLPPGPLKLPIIGNIHNLIGSLPHHRLRDLST 198
Query: 68 VYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYG 127
YGPLMHL+LGE+S IVVSS EYA+EVMK HD++FAS+P I A I+ Y S +AF+PYG
Sbjct: 199 KYGPLMHLKLGEVSTIVVSSAEYAKEVMKNHDLVFASRPPIQASKIMSYDSLGLAFAPYG 378
Query: 128 NYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTIT 187
+YWR LRKICT+ELL+ KRV SF+PIR EE TNLIK I S++GS IN T+ V ++I TIT
Sbjct: 379 DYWRNLRKICTLELLSSKRVQSFQPIRSEEVTNLIKWISSKEGSQINFTKEVFSTISTIT 558
Query: 188 TRAAFGNKCKGQEQLVSLA-NGESVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDL 246
+R AFG KCK ++ +S+ + + GGFD+G+L+PS + LQ +SG + K+E LH+Q DL
Sbjct: 559 SRTAFGKKCKENQKFISIVRDAIKIAGGFDLGDLYPSCRLLQNISGLKPKLEKLHKQADL 738
Query: 247 ILVNIINEHKEA-KSKAKEG---EVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILDI 302
I+ NII+EH+E KS+ E EVEEDLVDVL K G L DN++KA+ILD+
Sbjct: 739 IMQNIIDEHREVNKSRVNENQGEEVEEDLVDVLLKQDG---------LNDNSVKAVILDM 891
Query: 303 IGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVKE 362
G G ETS+TTI WAMAE++++P++M+K Q EVR++F+K+ E+ + +L YLK VVKE
Sbjct: 892 YGGGSETSATTITWAMAEMIKNPKIMEKVQAEVREVFDKERNPNESDMEKLTYLKYVVKE 1071
Query: 363 TLRLHPPTPLLL-RECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHP 416
TLRLHPP LL RECGQACEI GY IP KSKVIVNAW IGRDP +W +PERF+P
Sbjct: 1072TLRLHPPAAFLLPRECGQACEINGYDIPFKSKVIVNAWAIGRDPNHWDDPERFYP 1236
>TC90968 similar to SP|O81971|C7D9_SOYBN Cytochrome P450 71D9 (EC 1.14.-.-)
(P450 CP3). [Soybean] {Glycine max}, partial (64%)
Length = 1112
Score = 384 bits (987), Expect(2) = e-112
Identities = 203/354 (57%), Positives = 263/354 (73%), Gaps = 7/354 (1%)
Frame = +3
Query: 74 HLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYGNYWRQL 133
HL+LGE+S IVVSS EYA+EV+KTHD++FAS+P I A I+ Y S ++FSPYG+YWRQL
Sbjct: 3 HLKLGEVSTIVVSSAEYAKEVLKTHDLVFASRPPIQASKIMSYNSIGLSFSPYGDYWRQL 182
Query: 134 RKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTITTRAAFG 193
RKIC +ELL+ KRV SF+PIR EE TNLIK I S++GS IN+T+ V + I IT+R AFG
Sbjct: 183 RKICALELLSSKRVQSFQPIRSEEMTNLIKWIASKEGSEINLTKEVNSRIFLITSRVAFG 362
Query: 194 NKCKGQEQLVSLA-NGESVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDLILVNII 252
+CK ++ +SL V GGF++G+L+PS KWLQ +SG + K+E LH+Q D+IL NII
Sbjct: 363 KECKDNKKFISLVWEATRVAGGFNLGDLYPSYKWLQNISGLKPKLEKLHKQTDVILQNII 542
Query: 253 NEHKEA-KSKAKEG---EVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILDIIGAGGE 308
+EH+ KS+A E EV EDLVDVL K QD CL+DN++KA+ILD+ G G E
Sbjct: 543 DEHRLVDKSRAIEDHSEEVAEDLVDVLLK--------QD-CLSDNSVKAVILDMYGGGSE 695
Query: 309 TSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVKETLRLHP 368
TS++TI+WAMAE++++PR+MKK Q EVR++F K+ E+ + +L+YLK VVKETLRLHP
Sbjct: 696 TSASTILWAMAEMIKNPRIMKKLQAEVREVFEKERKPNESDMEKLKYLKCVVKETLRLHP 875
Query: 369 PTPLLL-RECGQACEIEGYHIPAKSKVIVNA-WTIGRDPKYWTEPERFHPERFI 420
P LL RECGQACEI GY IP KSKVIVN+ + W +PERF+PERFI
Sbjct: 876 PGAFLLPRECGQACEINGYGIPFKSKVIVNSLGPLEEIQIIWDDPERFYPERFI 1037
Score = 40.8 bits (94), Expect(2) = e-112
Identities = 15/18 (83%), Positives = 18/18 (99%)
Frame = +1
Query: 426 YKGNNFEYIPFGAGRRIC 443
YKGNNFE+IPFG+GRR+C
Sbjct: 1057 YKGNNFEFIPFGSGRRMC 1110
>TC87269 similar to PIR|T05940|T05940 cytochrome P450 83D1p - soybean
(fragment), partial (79%)
Length = 1878
Score = 397 bits (1019), Expect = e-111
Identities = 213/510 (41%), Positives = 316/510 (61%), Gaps = 20/510 (3%)
Frame = +1
Query: 8 LLALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAK 67
LL+L+ I ++ + K R+ + SS P PGP LP+IG++H L S+PH L L+K
Sbjct: 238 LLSLLPILILFIIHIYKIRSTSRASSTP---PGPKPLPLIGNLHQLDPSSPHHSLWKLSK 408
Query: 68 VYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYG 127
YGP+M LQLG I ++VSS + A +V+KTHD+ FAS+P + + L Y D+AF+PY
Sbjct: 409 HYGPIMSLQLGYIPTLIVSSAKMAEQVLKTHDLKFASRPSFLGLRKLSYNGLDLAFAPYS 588
Query: 128 NYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSP--INITQAVVTSIMT 185
YWR++RK+C L + +RV SFRP+RE E LI+ + G N+++ +++ T
Sbjct: 589 PYWREMRKLCVQHLFSSQRVHSFRPVRENEVAQLIQKLSQYGGDEKGANLSEILMSLTNT 768
Query: 186 ITTRAAFGNK--CKGQEQL------------VSLANGESVGGGFDIGELFPSAKWLQLVS 231
I + AFG C +E++ V L +++ F + FP W+ V
Sbjct: 769 IICKIAFGKTYVCDYEEEVELGSGQKRSRLQVLLNEAQALLAEFYFSDNFPLLGWIDRVK 948
Query: 232 GFRSKIEVLHRQIDLILVNIINEHKE--AKSKAKEGEVEEDLVDVLQKFQGGNDIDQDIC 289
G K++ +++DLI +I++H + A+ K KE EV +D++D+L + + + D
Sbjct: 949 GTLGKLDKTFKELDLIYQRVIDDHMDNSARPKTKEQEV-DDIIDILLQMMNDHSLSFD-- 1119
Query: 290 LTDNNIKAIILDIIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENY 349
LT ++IKA++++I AG +TSS +VWAM L+ +PRVM K Q E+R ++ K + E+
Sbjct: 1120LTLDHIKAVLMNIFIAGTDTSSAIVVWAMTTLMNNPRVMNKVQMEIRNLYEDKDFINEDD 1299
Query: 350 INELEYLKLVVKETLRLHPPTPLLL-RECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYW 408
I +L YLK VVKET+RL PP+PLL+ RE + C I+GY I K+ V VNAW I RDP+ W
Sbjct: 1300IEKLPYLKAVVKETIRLFPPSPLLVPRETIENCNIDGYEIKPKTLVYVNAWAIARDPENW 1479
Query: 409 TEPERFHPERFIGSSIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRL 468
+PE F+PERFI SS+D+KG NFE IPFG+GRR+CP + G++ VEL LA LL FDW L
Sbjct: 1480KDPEEFYPERFIISSVDFKGKNFELIPFGSGRRMCPAMNMGVVTVELTLANLLQSFDWNL 1659
Query: 469 PNGM-KGEDLDMTEQFGANVKRKSDLYLIP 497
P+G K + LD + G + +K DL+L+P
Sbjct: 1660PHGFDKEQVLDTQVKPGITMHKKIDLHLVP 1749
>TC78857 weakly similar to SP|O81974|C7D8_SOYBN Cytochrome P450 71D8 (EC
1.14.-.-) (P450 CP7). [Soybean] {Glycine max}, partial
(38%)
Length = 1724
Score = 346 bits (888), Expect = 1e-95
Identities = 187/495 (37%), Positives = 293/495 (58%), Gaps = 7/495 (1%)
Frame = +3
Query: 9 LALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAKV 68
L L++FF+FM K + K SS+ P GP LPIIG++H L +S H + +L+K+
Sbjct: 111 LPLLVFFLFM-----KYKTNIKNSSSSTFPKGPKGLPIIGNLHQLDTSNLHLQFWNLSKI 275
Query: 69 YGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYGN 128
YGPL LQ+G AIVV S + A+E++K HD +S+P L Y D+ FSPY +
Sbjct: 276 YGPLFSLQIGFKKAIVVCSSKLAQEILKDHDHDVSSRPPSHGPKTLSYNGIDMIFSPYND 455
Query: 129 YWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSP--INITQAVVTSIMTI 186
WR++RKIC + + K++SSF +R+ E +I+ I + S N+++ +++ +I
Sbjct: 456 CWREIRKICVVHFFSSKKISSFAHVRKSEVKLMIEKISNHVCSSKISNLSEVLMSVSSSI 635
Query: 187 TTRAAFGNKCKGQEQLVSLANG-----ESVGGGFDIGELFPSAKWLQLVSGFRSKIEVLH 241
R AFG + + S +G +++ F + + P W+ ++G ++++
Sbjct: 636 VCRIAFGKSYEHEGGEKSRFHGLLNETQAIFLSFFVSDYIPFMGWVDKLTGAIARVDNTF 815
Query: 242 RQIDLILVNIINEHKEAKSKAKEGEVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILD 301
+ +D ++ EH ++ K+ E E+D+VDVL + + + D LT+N+IKA++++
Sbjct: 816 KALDEFFEQVLKEHLNPNNRKKDDE-EKDIVDVLLELKNQGRLSID--LTNNHIKAVVMN 986
Query: 302 IIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVK 361
++ A +TS+ T VW M L+++PR MKKAQ E+R I KK + E+ I + Y K V+K
Sbjct: 987 LLVAATDTSAATSVWVMTGLIKNPRAMKKAQEEIRNI--KKEFIDEDDIQKFVYFKAVIK 1160
Query: 362 ETLRLHPPTPLLLRECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHPERFIG 421
ETLR + P PL RE ++ + GY I K+ V V+ W+I RDP+ W +P+ F+PERF+
Sbjct: 1161ETLRFYSPAPLAPRETSKSFTLNGYKIEPKTSVFVSIWSIHRDPETWKDPDEFYPERFLN 1340
Query: 422 SSIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMTE 481
+ ID+KG NFE+IPFGAGRRICPGI G+ VE+ A LL FDW +P GM ED+D
Sbjct: 1341NDIDFKGQNFEFIPFGAGRRICPGIPLGIATVEMITANLLNSFDWEMPEGMTKEDIDTEG 1520
Query: 482 QFGANVKRKSDLYLI 496
G +K+ L L+
Sbjct: 1521LPGLARHKKNHLCLV 1565
>TC80193 weakly similar to SP|O22307|C7DB_LOTJA Cytochrome P450 71D11 (EC
1.14.-.-) (Fragment). {Lotus japonicus}, partial (32%)
Length = 941
Score = 338 bits (866), Expect = 4e-93
Identities = 179/313 (57%), Positives = 233/313 (74%), Gaps = 4/313 (1%)
Frame = +2
Query: 12 VLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAKVYGP 71
+ FFI + +V+ K K + NLP GP KLPIIG+IH+L+SS PH+KLRDLA+ YGP
Sbjct: 8 IFFFILLAIVVQKVGKKLKNTDPFNLPNGPRKLPIIGNIHNLLSSQPHRKLRDLAREYGP 187
Query: 72 LMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYGNYWR 131
LMHLQLGEIS IV+SSP+ AREVMKTHDI FA++P+ ++ D++ Y ST I SPYGNYWR
Sbjct: 188 LMHLQLGEISFIVISSPDCAREVMKTHDINFATRPQGLSSDLIAYNSTGIVSSPYGNYWR 367
Query: 132 QLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTITTRAA 191
QLRKICT+ELL+ KRV+S++PIREEEF+NL+K I S++GSPIN+TQAV++SI TI +++A
Sbjct: 368 QLRKICTLELLSLKRVNSYQPIREEEFSNLVKWIASKEGSPINLTQAVLSSIYTIVSKSA 547
Query: 192 FGNKCKGQEQLVSLANGE--SVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDLILV 249
FG K K ++ + + FPS W+ +G R K+E +HR +D I+
Sbjct: 548 FGKKFKVPQKNLYQQKKRY*RLQQVLT*QSYFPSITWMHFFTGLRPKLERVHRVVDQIME 727
Query: 250 NIINEHKEAKSKAKEGEVE--EDLVDVLQKFQGGNDIDQDICLTDNNIKAIILDIIGAGG 307
NIINEHKEAKSK + +VE EDLVDVL K+Q GN +++ LT +NIKAII+DI GAGG
Sbjct: 728 NIINEHKEAKSKGEFDQVESDEDLVDVLLKYQDGN--NKEFFLTIDNIKAIIMDIFGAGG 901
Query: 308 ETSSTTIVWAMAE 320
ETS++TI WAMAE
Sbjct: 902 ETSASTIDWAMAE 940
>TC86485 similar to GP|23315211|gb|AAN20392.1 Sequence 405 from patent US
6410718, partial (90%)
Length = 1927
Score = 270 bits (690), Expect(2) = 7e-93
Identities = 149/420 (35%), Positives = 243/420 (57%), Gaps = 10/420 (2%)
Frame = +3
Query: 7 YLLALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLA 66
+L+A++ +L++ R LK+ P PPGP LPIIG++ ++ H+ L +LA
Sbjct: 183 FLMAIMFIVPLILLLGLVSRILKR----PRYPPGPIGLPIIGNML-MMDQLTHRGLANLA 347
Query: 67 KVYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPY 126
K YG + HL++G + + +S + AR+V++ D IF+++P VAI L Y D+AF+ Y
Sbjct: 348 KKYGGIFHLRMGFLHMVAISDADAARQVLQVQDNIFSNRPATVAIKYLTYDRADMAFAHY 527
Query: 127 GNYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTI 186
G +WRQ+RK+C ++L ++K S++ +R+E ++++ G+P+NI + V I
Sbjct: 528 GPFWRQMRKLCVMKLFSRKHAESWQSVRDE-VDYAVRTVSDNIGNPVNIGELVFNLTKNI 704
Query: 187 TTRAAFGNKCK-GQEQLVSLANGES-VGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQI 244
RAAFG+ + GQ++ + + S + G F+I + P + G +++ +++
Sbjct: 705 IYRAAFGSSSREGQDEFIGILQEFSKLFGAFNISDFVPCFGAID-PQGLNARLVKARKEL 881
Query: 245 DLILVNIINEHKEAKSKAKEGEVEEDLVDVLQKFQG--------GNDIDQDICLTDNNIK 296
D + II+EH + K + E D+VD L F +D++ I LT +NIK
Sbjct: 882 DSFIDKIIDEHVQKKKSVVDEET--DMVDELLAFYSEEAKLNNESDDLNNSIKLTKDNIK 1055
Query: 297 AIILDIIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYL 356
AII+D++ G ET ++ I WAMAEL++ P +KK Q E+ ++ V E+ +L YL
Sbjct: 1056AIIMDVMFGGTETVASAIEWAMAELMKSPEDLKKVQQELAEVVGLSRQVEESDFEKLTYL 1235
Query: 357 KLVVKETLRLHPPTPLLLRECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHP 416
K +KETLRLHPP PLLL E + + GY IP +++V++NAW IGRD W EP+ F P
Sbjct: 1236KCALKETLRLHPPIPLLLHETAEEATVNGYFIPKQARVMINAWAIGRDANCWEEPQSFKP 1415
Score = 89.4 bits (220), Expect(2) = 7e-93
Identities = 40/74 (54%), Positives = 54/74 (72%)
Frame = +1
Query: 425 DYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMTEQFG 484
D+KG+NFE+IPFG+GRR CPG+ GL ++LA+A LL+ F W LP+GMK ++DM++ FG
Sbjct: 1444 DFKGSNFEFIPFGSGRRSCPGMQLGLYALDLAVAHLLHCFTWELPDGMKPSEMDMSDVFG 1623
Query: 485 ANVKRKSDLYLIPT 498
R S L IPT
Sbjct: 1624 LTAPRASRLVAIPT 1665
>TC85609 weakly similar to SP|Q9LTL2|C72P_ARATH Cytochrome P450 71B25 (EC
1.14.-.-). [Mouse-ear cress] {Arabidopsis thaliana},
partial (33%)
Length = 1786
Score = 336 bits (862), Expect = 1e-92
Identities = 197/504 (39%), Positives = 292/504 (57%), Gaps = 10/504 (1%)
Frame = +1
Query: 3 SPIFYL-LALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQK 61
SP+ L AL+LFF+F K KKS+ LPPGP LP IG++H L SS
Sbjct: 58 SPLILLPFALLLFFLF------KKHKTSKKSTT--LPPGPKGLPFIGNLHQLDSSVLGLN 213
Query: 62 LRDLAKVYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDI 121
+L+K YGP++ L+LG +VVSS + A+EVMKTHDI F ++P +++ + Y D
Sbjct: 214 FYELSKKYGPIISLKLGSKQTVVVSSAKMAKEVMKTHDIEFCNRPALISHMKISYNGLDQ 393
Query: 122 AFSPYGNYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSP--INITQAV 179
F+PY YWR +K+ I L+ KRVS F +R++E T +IK I S +N+ +
Sbjct: 394 IFAPYREYWRHTKKLSFIHFLSVKRVSMFYSVRKDEVTRMIKKISENASSNKVMNMQDLL 573
Query: 180 VTSIMTITTRAAFGNKCKGQEQLVSLANG-----ESVGGGFDIGELFPSAKWL-QLVSGF 233
T+ + AFG + +G+ S+ G + + F + P + ++G
Sbjct: 574 TCLTSTLVCKTAFGRRYEGEGIERSMFQGLHKEVQDLLISFFYADYLPFVGGIVDKLTGK 753
Query: 234 RSKIEVLHRQIDLILVNIINEHKEAKSKAKEGEVEEDLVDVLQKFQGGNDIDQDICLTDN 293
S++E + D + +I++EH + + K K E+D++D L + + ND + LT
Sbjct: 754 TSRLEKTFKVSDELYQSIVDEHLDPERK-KLPPHEDDVIDALIELK--NDPYCSMDLTAE 924
Query: 294 NIKAIILDIIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINEL 353
+IK +I+++ A ET + +VWAM L+++PR M+K Q E+RK+ KG + E + +L
Sbjct: 925 HIKPLIMNMSFAVTETIAAAVVWAMTALMKNPRAMQKVQEEIRKVCAGKGFIEEEDVEKL 1104
Query: 354 EYLKLVVKETLRLHPPTPLLL-RECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPE 412
Y K V+KE++RL+P P+LL RE C I GY IP K+ V VNA I RDP+ W +PE
Sbjct: 1105PYFKAVIKESMRLYPILPILLPRETMTNCNIAGYDIPDKTLVYVNALAIHRDPEVWKDPE 1284
Query: 413 RFHPERFIGSSIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRLPNGM 472
F+PERFIGS ID KG +FE IPFG+GRRICPG+ + ++L L+ LLY FDW +P G
Sbjct: 1285EFYPERFIGSDIDLKGQDFELIPFGSGRRICPGLNMAIATIDLVLSNLLYSFDWEMPEGA 1464
Query: 473 KGEDLDMTEQFGANVKRKSDLYLI 496
K ED+D Q G +K+ L L+
Sbjct: 1465KREDIDTHGQAGLIQHKKNPLCLV 1536
>TC77326 similar to GP|12004682|gb|AAG44132.1 cytochrome P450 {Pisum
sativum}, complete
Length = 1864
Score = 335 bits (860), Expect = 2e-92
Identities = 183/496 (36%), Positives = 285/496 (56%), Gaps = 10/496 (2%)
Frame = +1
Query: 11 LVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAKVYG 70
+ LF F++++L R + K NLPPGP PIIG+++ L+ S PHQ L L + YG
Sbjct: 88 VTLFATFVILLLFSRRLRRHKY---NLPPGPKPWPIIGNLN-LIGSLPHQSLHGLTQKYG 255
Query: 71 PLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYGNYW 130
P+MHL G IV ++ E A+ +KTHD A +PK+ A Y +DI +S YG YW
Sbjct: 256 PIMHLYFGSKPVIVGATVELAKSFLKTHDATLAGRPKLSAGKYTTYNYSDITWSQYGPYW 435
Query: 131 RQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTITTRA 190
RQ R++C +EL + KR+ S+ IR++E + + + + + I + + T + + +R
Sbjct: 436 RQARRMCLLELFSAKRLESYEYIRKQEMHDFLHKLFNSKNKTILVKDHLSTLSLNVISRM 615
Query: 191 AFGNKCKGQEQLVSLANGES---------VGGGFDIGELFPSAKWLQLVSGFRSKIEVLH 241
G K + ++ E + G +IG+ P +L L G+ +++ L
Sbjct: 616 VLGKKYLEKTDNAVISPDEFKKMLDELFLLNGILNIGDFIPWIHFLDL-QGYVKRMKTLS 792
Query: 242 RQIDLILVNIINEHKEAKSKAKEGEVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILD 301
++ D + +++ EH E + K+ V +D+VDVL Q D + ++ L + +KA D
Sbjct: 793 KKFDRFMEHVLEEHIERRKNVKD-YVAKDMVDVL--LQLAEDPNLEVKLERHGVKAFTQD 963
Query: 302 IIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVK 361
+I G E+S+ T+ WA++ELVR P + KKA E+ ++ K V E I L Y+ + K
Sbjct: 964 LIAGGTESSAVTVEWAISELVRKPEIFKKATEELDRVIGKDRWVEEKDIANLPYVYAIAK 1143
Query: 362 ETLRLHPPTPLLL-RECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHPERFI 420
ET+RLHP P L+ RE + C+++GY IP + V+VN WTI RD + W P F PERF+
Sbjct: 1144ETMRLHPVAPFLVPREAREDCKVDGYDIPKGTIVLVNTWTIARDSEVWENPYEFMPERFL 1323
Query: 421 GSSIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMT 480
G ID KG++FE +PFGAGRR+CPG G+ ++ +LA LL+ F+W LPN +K EDL+M
Sbjct: 1324GKDIDVKGHDFELLPFGAGRRMCPGYPLGIKVIQTSLANLLHGFNWTLPNNVKKEDLNME 1503
Query: 481 EQFGANVKRKSDLYLI 496
E FG + +K L ++
Sbjct: 1504EIFGLSTPKKIPLEIV 1551
>TC90108 similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (EC
1.14.-.-). [Soybean] {Glycine max}, partial (41%)
Length = 958
Score = 332 bits (851), Expect = 2e-91
Identities = 173/299 (57%), Positives = 222/299 (73%), Gaps = 1/299 (0%)
Frame = +1
Query: 200 EQLVSLANGESVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDLILVNIINEHKEAK 259
E++VSL GG I +L+PS K LQ VS ++K+E LH++ID+IL +II++HK
Sbjct: 16 EEIVSLL------GGICIVDLYPSIKLLQCVSRAKAKMEKLHKEIDMILQDIIDDHK--- 168
Query: 260 SKAKEGEVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILDIIGAGGETSSTTIVWAMA 319
S KE EE LVDVL K Q N Q LTD+NIK+II DI AG ETSSTT++WA++
Sbjct: 169 SIHKEDSNEEYLVDVLLKIQQENYHSQHP-LTDDNIKSIIQDIFDAGTETSSTTVLWAIS 345
Query: 320 ELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVKETLRLHPPTPLLL-RECG 378
E+V++P+VM++AQ EVR++F++KG V E +++L YLK V+KET+RLHP PLLL RE
Sbjct: 346 EMVKNPKVMEEAQAEVRRVFDRKGFVDETELHQLIYLKSVIKETMRLHPTVPLLLPRESR 525
Query: 379 QACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHPERFIGSSIDYKGNNFEYIPFGA 438
+ C+I GY IPAK++V+VNAW IGRDP+YW + E F PERF+ S ID+KG +FEYIPFGA
Sbjct: 526 ERCQINGYEIPAKTRVMVNAWAIGRDPRYWVDAESFKPERFVNSPIDFKGTDFEYIPFGA 705
Query: 439 GRRICPGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMTEQFGANVKRKSDLYLIP 497
GR++CPGI F L NVEL LA LLYHFDW+LPN MK E+LDMTE FG RK +L LIP
Sbjct: 706 GRKMCPGIAFALPNVELPLASLLYHFDWKLPNKMKNEELDMTESFGITAGRKHNLCLIP 882
>TC82354 similar to SP|O22307|C7DB_LOTJA Cytochrome P450 71D11 (EC 1.14.-.-)
(Fragment). {Lotus japonicus}, partial (43%)
Length = 919
Score = 328 bits (842), Expect = 2e-90
Identities = 157/243 (64%), Positives = 196/243 (80%), Gaps = 3/243 (1%)
Frame = +1
Query: 263 KEG--EVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILDIIGAGGETSSTTIVWAMAE 320
KEG EV+EDL+D L KF+ D + LT +NIKAIILD+ AG ETS++TI+WAMAE
Sbjct: 1 KEGLAEVQEDLIDCLLKFEDSGS-DMGLNLTTDNIKAIILDVFAAGSETSASTIIWAMAE 177
Query: 321 LVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVKETLRLHPPTPLLL-RECGQ 379
+++D RV+KKAQ EVR+ F++ G V E I+E +YLK V+KE+LRLHP PLLL RECGQ
Sbjct: 178 MMKDQRVLKKAQAEVREGFDRSGRVDEATIDEFKYLKAVIKESLRLHPSVPLLLPRECGQ 357
Query: 380 ACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHPERFIGSSIDYKGNNFEYIPFGAG 439
ACEI GY+IP KS+V+VNAW IGRDPKYW +P++F+PERFI SSID+KG NFEYIPFGAG
Sbjct: 358 ACEINGYYIPVKSRVLVNAWAIGRDPKYWNDPDKFYPERFIDSSIDFKGTNFEYIPFGAG 537
Query: 440 RRICPGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMTEQFGANVKRKSDLYLIPTI 499
RRICPG+ +G+ NVE LAL+LYHFDW+LPNG+K E+L++ E+FGA + RK DLYLIP I
Sbjct: 538 RRICPGMNYGMANVEQVLALILYHFDWKLPNGIKNEELELIEEFGAAMSRKGDLYLIPII 717
Query: 500 PLP 502
P
Sbjct: 718 SHP 726
>TC77258 similar to GP|5059126|gb|AAD38930.1| cytochrome P450
monooxygenaseCYP93D1 {Glycine max}, partial (95%)
Length = 1823
Score = 326 bits (835), Expect = 1e-89
Identities = 185/491 (37%), Positives = 286/491 (57%), Gaps = 16/491 (3%)
Frame = +1
Query: 7 YLLALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGP-WKLPIIGHIHHLVSSTPHQKLRDL 65
Y++ +L+FI + + R+L KKS LPPGP PI+GH +L S H+ L L
Sbjct: 97 YVVLFLLWFISSIFI----RSLFKKSVCYKLPPGPPISFPILGHAPYL-RSLLHKSLYKL 261
Query: 66 AKVYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSP 125
+ YGPLMH+ LG +V S+ E A++++KT + F ++P ++A + L YG+ D F P
Sbjct: 262 SNRYGPLMHIMLGSQHVVVASTAESAKQILKTCEESFTNRPIMIASENLTYGAADYFFIP 441
Query: 126 YGNYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSI--DSQQGSPINITQAVVTSI 183
YGNYWR L+K+C ELL+ K + F IRE+E + ++ S+ G PI + ++
Sbjct: 442 YGNYWRFLKKLCMTELLSGKTLEHFVHIREDEIKCFMGTLLEISKNGKPIEMRHELIRHT 621
Query: 184 MTITTRAAFGNKCKGQEQLVSLAN------GESVGGGFDIGELFPSAKWLQLVSGFRSKI 237
I +R G K G V GE +G F++G++ + L L GF +
Sbjct: 622 NNIISRMTMGKKSSGMNDEVGQLRKVVREIGELLGA-FNLGDIIGFMRPLDL-QGFGKRN 795
Query: 238 EVLHRQIDLILVNIINEHKEAKSKAKEG-EVEEDLVDVLQKFQGGNDIDQDICLTDNNIK 296
+ H ++D+++ ++ EH+EA++K G + ++DL D+L +D + LT + K
Sbjct: 796 KDTHHKMDVMMEKVLKEHEEARAKEGAGSDRKKDLFDILLNLIEADD-GAESKLTRQSAK 972
Query: 297 AIILDIIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYL 356
A LD+ AG ++ + WA+AEL+R+P V KKA+ E+ K+ E+ I L YL
Sbjct: 973 AFALDMFIAGTNGPASVLEWALAELIRNPHVFKKAREEIDSTVGKERLFKESDIPNLPYL 1152
Query: 357 KLVVKETLRLHPPTPLLLRECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHP 416
+ VVKETLR+HPPTP+ RE ++C+++GY +PA SK+ +NAW IGRDP YW P F+P
Sbjct: 1153QAVVKETLRMHPPTPIFAREATRSCQVDGYDVPAFSKIFINAWAIGRDPNYWDNPLVFNP 1332
Query: 417 ERFI------GSSIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRLPN 470
ERF+ S ID +G ++ +PFG+GRR CPG + L+ ++ LA L+ FDW + +
Sbjct: 1333ERFLQSDDPSKSKIDVRGQYYQLLPFGSGRRSCPGSSLALLVIQATLASLIQCFDWVVND 1512
Query: 471 GMKGEDLDMTE 481
G K D+DM+E
Sbjct: 1513G-KSHDIDMSE 1542
>TC85610 similar to GP|21068674|emb|CAD31843. putative cytochrome P450
monooxygenase {Cicer arietinum}, complete
Length = 1578
Score = 223 bits (567), Expect(2) = 6e-89
Identities = 142/400 (35%), Positives = 221/400 (54%), Gaps = 10/400 (2%)
Frame = +2
Query: 3 SPIFYL-LALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQK 61
SP+ L AL+LFF+F K KKS+ LPPGP LP IG++H L SS
Sbjct: 11 SPLILLPFALLLFFLF------KKHKTSKKSTT--LPPGPKGLPFIGNLHQLDSSALGLN 166
Query: 62 LRDLAKVYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDI 121
+L+K YG L++L+LG IVVSS + A++VMKTHDI F ++P +++ Y D
Sbjct: 167 FYELSKKYGSLIYLKLGSRQTIVVSSAKMAKQVMKTHDIDFCNRPALISHMKFSYDGLDQ 346
Query: 122 AFSPYGNYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGS--PINITQAV 179
FSPY YWR +K+ I L+ KRV F +R+ E T +I I S +N+ + +
Sbjct: 347 FFSPYREYWRHTKKLSFIHFLSVKRVVMFSSVRKYETTQMITKISEHASSNKVVNLHELL 526
Query: 180 VTSIMTITTRAAFGNKCKGQEQLVS-----LANGESVGGGFDIGELFPSAKWL-QLVSGF 233
+ + R AFG + + + S L + + F + P + +G
Sbjct: 527 MCLTSAVVCRTAFGRRFEDEAAERSMFHDLLKEAQEMTISFFYTDYLPFVGGIVDKFTGL 706
Query: 234 RSKIEVLHRQIDLILVNIINEHKEAKSKAKEGEVEEDLVDVLQKFQGGNDIDQDICLTDN 293
S++E L + +D ++ +EH + K K EED++D L + + ND + L+
Sbjct: 707 MSRLEKLFKILDGFFQSVFDEHIDPNRK-KLPPHEEDVIDALIELK--NDPYCSMDLSAE 877
Query: 294 NIKAIILDIIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINEL 353
+IK +I++++ AG +T + +VWAM L+++PRVM+K Q E+RK + KG + E + +L
Sbjct: 878 HIKPLIMNMLLAGTDTIAAAVVWAMTALMKNPRVMQKVQEEIRKAYEGKGFIEEEDVQKL 1057
Query: 354 EYLKLVVKETLRLHPPTPLLL-RECGQACEIEGYHIPAKS 392
Y K V+KE++RL+P P+LL RE + C+IEGY IP K+
Sbjct: 1058PYFKAVIKESMRLYPSLPVLLPRETMKKCDIEGYEIPDKN 1177
Score = 123 bits (309), Expect(2) = 6e-89
Identities = 56/113 (49%), Positives = 76/113 (66%)
Frame = +3
Query: 384 EGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHPERFIGSSIDYKGNNFEYIPFGAGRRIC 443
+G ++ V +NAW I RDP+ W +PE F+PERFIGS ID KG +FE IPFG+GRR+C
Sbjct: 1152 KGTKFQTRTLVYINAWAIHRDPEAWKDPEEFYPERFIGSDIDLKGQDFELIPFGSGRRVC 1331
Query: 444 PGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMTEQFGANVKRKSDLYLI 496
PG+ + V+L LA LLY FDW +P G+K E++D+ G +K+ L LI
Sbjct: 1332 PGLNMAIATVDLVLANLLYLFDWEMPEGVKWENIDIDGLPGLVQHKKNPLCLI 1490
>TC79360 similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (EC
1.14.-.-). [Soybean] {Glycine max}, partial (52%)
Length = 1134
Score = 323 bits (829), Expect = 7e-89
Identities = 169/306 (55%), Positives = 215/306 (70%), Gaps = 3/306 (0%)
Frame = +1
Query: 200 EQLVSLANGESVG--GGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDLILVNIINEHKE 257
+QL E++G G F I +L+PS K LQ VS ++++E L +ID IL +IIN+H+
Sbjct: 1 QQLFKSTIEEALGLLGEFCIADLYPSIKILQKVSRVKTRVERLQGEIDRILQDIINDHRN 180
Query: 258 AKSKAKEGEVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILDIIGAGGETSSTTIVWA 317
SK + +EDLVDVL K Q N Q LTD NIK++I D+ AG ETSS ++WA
Sbjct: 181 NHSKTSK---DEDLVDVLLKVQHENVHSQQP-LTDENIKSVIQDLFIAGSETSSGIVLWA 348
Query: 318 MAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVKETLRLHPPTPLLL-RE 376
M+E++++P VM++AQ EVR++F+KKG V E + +L YLK V+KET RLHP PLL+ RE
Sbjct: 349 MSEMIKNPIVMEEAQVEVRRVFDKKGYVDETELQQLTYLKCVIKETFRLHPTVPLLVPRE 528
Query: 377 CGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHPERFIGSSIDYKGNNFEYIPF 436
+ CEI GY IPAK++V VN W IGRDPKYW E E F PERF+ SSID+KG +FE IPF
Sbjct: 529 SRERCEINGYEIPAKTRVAVNVWAIGRDPKYWVEAESFKPERFVNSSIDFKGTDFELIPF 708
Query: 437 GAGRRICPGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMTEQFGANVKRKSDLYLI 496
GAGRR+CPGI F L NVEL LA LLYHFDW+LPNGM ++LDMTE FG V +K D+ LI
Sbjct: 709 GAGRRMCPGIAFALPNVELPLAKLLYHFDWKLPNGMSHQELDMTESFGLTVGKKHDVCLI 888
Query: 497 PTIPLP 502
P P
Sbjct: 889 PITRRP 906
>AW775042 weakly similar to SP|O22307|C7DB Cytochrome P450 71D11 (EC
1.14.-.-) (Fragment). {Lotus japonicus}, partial (29%)
Length = 699
Score = 312 bits (799), Expect = 2e-85
Identities = 152/232 (65%), Positives = 189/232 (80%), Gaps = 1/232 (0%)
Frame = +1
Query: 25 GRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAKVYGPLMHLQLGEISAIV 84
G+ LKK S NLP GP KLPIIG+IH+L+SS PH+KL DLAK YGPLMHLQLGE+S IV
Sbjct: 4 GKKLKKTDSTFNLPKGPRKLPIIGNIHNLLSSQPHRKLSDLAKKYGPLMHLQLGEVSTIV 183
Query: 85 VSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYGNYWRQLRKICTIELLTQ 144
+SS E+A+EVMKTHDI FA++PK++AIDI+ Y ST I+FSPYG+YWRQ RKICT+ELL+
Sbjct: 184 ISSAEFAKEVMKTHDIHFATRPKVLAIDIMSYNSTSISFSPYGDYWRQARKICTLELLSL 363
Query: 145 KRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTITTRAAFGNKCKGQEQLVS 204
KRV+S++PIREE F+NLIKSI S+ GSPIN T+AV++SI TI +RAAFGNKCK QE+ +S
Sbjct: 364 KRVNSYQPIREEVFSNLIKSIASENGSPINFTEAVLSSIYTIVSRAAFGNKCKDQEKFIS 543
Query: 205 LA-NGESVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDLILVNIINEH 255
+ + GFD+G+ FPS+KWLQ V+G R KIE H+Q D IL NII+EH
Sbjct: 544 VVKESVKIAAGFDLGDSFPSSKWLQYVTGLRPKIEKFHQQTDEILENIIDEH 699
>BG586162 weakly similar to SP|O22307|C7DB Cytochrome P450 71D11 (EC
1.14.-.-) (Fragment). {Lotus japonicus}, partial (42%)
Length = 778
Score = 307 bits (787), Expect(2) = 8e-85
Identities = 154/237 (64%), Positives = 191/237 (79%), Gaps = 1/237 (0%)
Frame = +1
Query: 12 VLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVSSTPHQKLRDLAKVYGP 71
+ FFI + +++ K R KK SA N+PPGP KLPIIG+IH+L+SS PHQKLRDLAK YGP
Sbjct: 4 IFFFILLTLIVQKIRK-KKSYSAYNIPPGPRKLPIIGNIHNLLSSQPHQKLRDLAKKYGP 180
Query: 72 LMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYGNYWR 131
+MHLQLGE+SA+V+SSPE A EVMKTHDI F+S+P+I+A +I+ Y ST IAF+PYGNYWR
Sbjct: 181 VMHLQLGEVSAMVISSPECASEVMKTHDIHFSSRPQILATEIMSYNSTSIAFAPYGNYWR 360
Query: 132 QLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTITTRAA 191
QLRKICT+ELL+ KRV+S++PIREE F NL K I SQ G PINIT+AV +SI TI +RAA
Sbjct: 361 QLRKICTLELLSLKRVNSYQPIREEVFFNLAKWIASQVGIPINITEAVRSSIYTIVSRAA 540
Query: 192 FGNKCKGQEQLVSLANGE-SVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDLI 247
FGN+CK Q++ +S+ V GGFD+G+LFPSAKWLQLV+G R K+E HRQ D I
Sbjct: 541 FGNECKDQDKFISVVKQSIKVAGGFDLGDLFPSAKWLQLVTGMRPKLERFHRQTDQI 711
Score = 25.0 bits (53), Expect(2) = 8e-85
Identities = 12/18 (66%), Positives = 15/18 (82%), Gaps = 1/18 (5%)
Frame = +3
Query: 250 NIINEHKEAK-SKAKEGE 266
NIINEHKE K +KAK+ +
Sbjct: 717 NIINEHKEEKYTKAKDDQ 770
>TC89202 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (EC
1.14.-.-). [Soybean] {Glycine max}, partial (38%)
Length = 1040
Score = 305 bits (781), Expect = 3e-83
Identities = 161/313 (51%), Positives = 222/313 (70%), Gaps = 2/313 (0%)
Frame = +1
Query: 33 SAPNLPPGPWKLPIIGHIHHLVS-STPHQKLRDLAKVYGPLMHLQLGEISAIVVSSPEYA 91
S NLPPGPWKLP IG++H ++S S PH + + LA YGPLM+L+LG + ++VSSPE A
Sbjct: 109 STVNLPPGPWKLPFIGNLHQIISRSLPHHRFKFLADKYGPLMYLKLGGVPYVIVSSPEIA 288
Query: 92 REVMKTHDIIFASKPKIVAIDILLYGSTDIAFSPYGNYWRQLRKICTIELLTQKRVSSFR 151
+E+MKTHD+ F+ +P ++ I Y +TD+ FS YG WRQLRKIC IELL+ KRV SFR
Sbjct: 289 KEIMKTHDLNFSGRPNLLLSTIWSYNATDVIFSIYGERWRQLRKICVIELLSAKRVQSFR 468
Query: 152 PIREEEFTNLIKSIDSQQGSPINITQAVVTSIMTITTRAAFGNKCKGQEQLVS-LANGES 210
IRE+E TNL+KSI + +GS +N+TQ ++++ IT RAAFG + K QE S + S
Sbjct: 469 SIREDEVTNLVKSITASEGSVVNLTQKILSTTYGITARAAFGKRSKHQEVFRSAIEEVAS 648
Query: 211 VGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQIDLILVNIINEHKEAKSKAKEGEVEED 270
+ GG I +LFPS K LQ +S ++K+E LH+++D+ L +II++H KS KE +ED
Sbjct: 649 LLGGVCIVDLFPSIKLLQCLSRAKTKMEKLHKELDMTLQDIIDDH---KSIHKEESNDED 819
Query: 271 LVDVLQKFQGGNDIDQDICLTDNNIKAIILDIIGAGGETSSTTIVWAMAELVRDPRVMKK 330
LVDVL K Q N Q LTD+NIK+II D+ GAG ETSS ++WAM+ +V++ VM++
Sbjct: 820 LVDVLLKIQQENYHSQH-PLTDDNIKSIIQDMFGAGTETSSEAVIWAMSAMVKNSNVMEQ 996
Query: 331 AQYEVRKIFNKKG 343
AQ EVR++F+KKG
Sbjct: 997 AQAEVRRVFDKKG 1035
>TC83902 similar to SP|O81974|C7D8_SOYBN Cytochrome P450 71D8 (EC 1.14.-.-)
(P450 CP7). [Soybean] {Glycine max}, partial (41%)
Length = 900
Score = 301 bits (772), Expect = 3e-82
Identities = 149/271 (54%), Positives = 204/271 (74%), Gaps = 5/271 (1%)
Frame = +1
Query: 234 RSKIEVLHRQIDLILVNIINEHKEAKSKAKE---GEVE-EDLVDVLQKFQGGNDIDQDIC 289
+ K+E +H+++ I NII +H+E + +AKE EV+ EDL+DVL + Q +++D I
Sbjct: 4 KPKLEKIHKRVF*IFENIIRQHQEKRERAKEDDNNEVDNEDLLDVLLRVQQSDNLD--IK 177
Query: 290 LTDNNIKAIILDIIGAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENY 349
+T NNIKA+I D+ AG +T+STTI WAM+E++++P V +KAQ E+R+ F K + E+
Sbjct: 178 ITTNNIKAVIWDVFVAGTDTTSTTIEWAMSEMMKNPSVREKAQAELREAFKGKKIISESD 357
Query: 350 INELEYLKLVVKETLRLHPPTPLLL-RECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYW 408
+NEL Y KLV+KET+RLHPP+PLL+ REC + I+GY IP +KV++NAW + RDP+YW
Sbjct: 358 LNELTYFKLVIKETMRLHPPSPLLVPRECTELTIIDGYEIPKNTKVMINAWAVARDPQYW 537
Query: 409 TEPERFHPERFIGSSIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRL 468
T+ E F PERF GS ID+KGNNFEYIPFGAGRR+CPG++FG+ +V L LALLLYHF+W L
Sbjct: 538 TDAEMFIPERFDGSLIDFKGNNFEYIPFGAGRRMCPGMSFGIASVMLPLALLLYHFNWEL 717
Query: 469 PNGMKGEDLDMTEQFGANVKRKSDLYLIPTI 499
PN MK EDLDMTE G V R+++L LIP +
Sbjct: 718 PNQMKPEDLDMTENVGLAVGRENELCLIPNV 810
>TC80192 weakly similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (EC
1.14.-.-). [Soybean] {Glycine max}, partial (35%)
Length = 829
Score = 301 bits (770), Expect = 5e-82
Identities = 142/200 (71%), Positives = 167/200 (83%), Gaps = 1/200 (0%)
Frame = +1
Query: 304 GAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNKKGTVGENYINELEYLKLVVKET 363
GAGG+TS+ TI WAMAE++R+PR+MKKAQ EVR++F G V ENYI+EL YLK VVKET
Sbjct: 1 GAGGDTSAITIDWAMAEMIREPRIMKKAQAEVREVFKLNGRVDENYISELNYLKSVVKET 180
Query: 364 LRLHPPTPLLL-RECGQACEIEGYHIPAKSKVIVNAWTIGRDPKYWTEPERFHPERFIGS 422
LRLHPP PLLL RECG+ACEI GY+IP KSKVIVNAW I RDP YW EPERF+PERF S
Sbjct: 181 LRLHPPGPLLLPRECGKACEINGYNIPFKSKVIVNAWAIARDPNYWAEPERFYPERFTDS 360
Query: 423 SIDYKGNNFEYIPFGAGRRICPGITFGLINVELALALLLYHFDWRLPNGMKGEDLDMTEQ 482
+ID KG+NFEYIPFGAGRRICPG FGL +VE+ALA+LLYHFDW+LP G+K E+LDMTE+
Sbjct: 361 TIDSKGDNFEYIPFGAGRRICPGSIFGLRSVEVALAMLLYHFDWKLPCGIKSEELDMTEE 540
Query: 483 FGANVKRKSDLYLIPTIPLP 502
FG ++RK DL L P++ P
Sbjct: 541 FGVTMRRKDDLVLFPSVYHP 600
>TC80864 similar to SP|O48923|C7DA_SOYBN Cytochrome P450 71D10 (EC
1.14.-.-). [Soybean] {Glycine max}, partial (46%)
Length = 1083
Score = 300 bits (769), Expect = 7e-82
Identities = 157/338 (46%), Positives = 233/338 (68%), Gaps = 2/338 (0%)
Frame = +3
Query: 6 FYLLALVLFFIFMLVVLTKGRNLKKKSSAPNLPPGPWKLPIIGHIHHLVS-STPHQKLRD 64
F + +L F+ +LV+ + +S LPPGPWKLP+IG++H ++S S PH +
Sbjct: 81 FSNIIFILSFLILLVLFKIVQRWSFNNSTTKLPPGPWKLPLIGNLHQIISRSLPHHLFKI 260
Query: 65 LAKVYGPLMHLQLGEISAIVVSSPEYAREVMKTHDIIFASKPKIVAIDILLYGSTDIAFS 124
LA YGPLMHL+LGE+ ++VSSPE A+E+MKTHD+ F +P ++ +I Y +TDIAF+
Sbjct: 261 LADKYGPLMHLKLGEVPYVIVSSPEIAKEIMKTHDLNFCDRPNLLLSNIYSYNATDIAFA 440
Query: 125 PYGNYWRQLRKICTIELLTQKRVSSFRPIREEEFTNLIKSIDSQQGSPINITQAVVTSIM 184
YG +WRQLRKIC IELL+ KRV SFR IRE+E +NL+KSI + +GS +N+T+ + +
Sbjct: 441 AYGEHWRQLRKICVIELLSAKRVQSFRSIREDEVSNLVKSITASEGSVVNLTRKIFSMTN 620
Query: 185 TITTRAAFGNKCKGQEQLV-SLANGESVGGGFDIGELFPSAKWLQLVSGFRSKIEVLHRQ 243
IT RAAFG + + Q+ + +L + G F+I +L+PS K LQ ++ ++K+E LH +
Sbjct: 621 GITARAAFGKRNRHQDVFIPALEKVVVLLGRFEIADLYPSIKLLQWMTREKTKMEKLHTE 800
Query: 244 IDLILVNIINEHKEAKSKAKEGEVEEDLVDVLQKFQGGNDIDQDICLTDNNIKAIILDII 303
ID+I +II++H +S KE +EDLVDVL K Q N + LTD+N+K+II D+
Sbjct: 801 IDMIAQDIIDDH---RSIHKEASNDEDLVDVLLKIQQEN-YHSEHPLTDDNMKSIIQDMF 968
Query: 304 GAGGETSSTTIVWAMAELVRDPRVMKKAQYEVRKIFNK 341
AG ETSS ++WAM+E+V++P+VM++AQ EV ++F+K
Sbjct: 969 LAGTETSSQVLLWAMSEMVKNPKVMEEAQDEVSRVFDK 1082
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.140 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,161,412
Number of Sequences: 36976
Number of extensions: 240006
Number of successful extensions: 1716
Number of sequences better than 10.0: 269
Number of HSP's better than 10.0 without gapping: 1449
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1460
length of query: 505
length of database: 9,014,727
effective HSP length: 100
effective length of query: 405
effective length of database: 5,317,127
effective search space: 2153436435
effective search space used: 2153436435
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140774.1


