

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.4 + phase: 0
(101 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
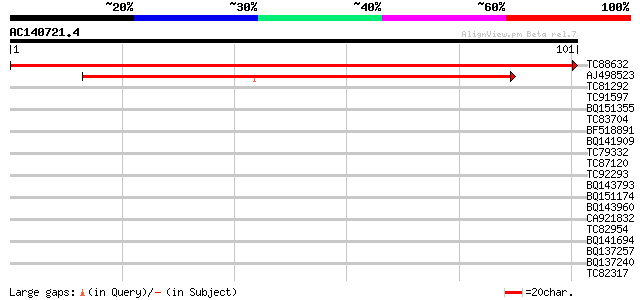
TC88632 homologue to GP|1754979|gb|AAB39474.1| Em protein {Robin... 203 8e-54
AJ498523 homologue to GP|1754977|gb| Em protein {Robinia pseudoa... 124 8e-30
TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalf... 30 0.11
TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protei... 30 0.11
BQ151355 homologue to GP|12620658|gb| ID556 {Bradyrhizobium japo... 29 0.33
TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19... 29 0.33
BF518891 weakly similar to GP|11993492|g ubiquitin-specific prot... 28 0.44
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 28 0.44
TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog... 28 0.74
TC87120 similar to SP|P48724|IF5_PHAVU Eukaryotic translation in... 28 0.74
TC92293 similar to SP|Q64733|FXB2_MOUSE Forkhead box protein B2 ... 27 0.97
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 27 0.97
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 27 0.97
BQ143960 weakly similar to SP|P21997|SSGP_ Sulfated surface glyc... 27 1.3
CA921832 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidops... 27 1.3
TC82954 weakly similar to GP|8809591|dbj|BAA97142.1 emb|CAB82814... 27 1.3
BQ141694 similar to GP|12697907|db KIAA1681 protein {Homo sapien... 27 1.3
BQ137257 similar to GP|21536912|gb| unknown {Arabidopsis thalian... 27 1.3
BQ137240 similar to PIR|F75311|F753 ABC transporter ATP-binding... 27 1.7
TC82317 similar to GP|2921209|gb|AAC04691.1| beta-ketoacyl-ACP s... 26 2.2
>TC88632 homologue to GP|1754979|gb|AAB39474.1| Em protein {Robinia
pseudoacacia}, partial (92%)
Length = 637
Score = 203 bits (517), Expect = 8e-54
Identities = 101/101 (100%), Positives = 101/101 (100%)
Frame = +3
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM
Sbjct: 78 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 257
Query: 61 GRKGGLSTMEKSGGERAEEEGIDIDESKFKTGGGGGRSQNK 101
GRKGGLSTMEKSGGERAEEEGIDIDESKFKTGGGGGRSQNK
Sbjct: 258 GRKGGLSTMEKSGGERAEEEGIDIDESKFKTGGGGGRSQNK 380
>AJ498523 homologue to GP|1754977|gb| Em protein {Robinia pseudoacacia},
partial (98%)
Length = 398
Score = 124 bits (310), Expect = 8e-30
Identities = 64/97 (65%), Positives = 72/97 (73%), Gaps = 20/97 (20%)
Frame = +2
Query: 14 KAKQGETVVPGGTGGKSLEAQEHLAEGRS--------------------KGGQTRKEQLG 53
+ + GETV+PGGTGGKSLEAQ+HLAEGRS KGGQTRKEQ+G
Sbjct: 77 EGRLGETVIPGGTGGKSLEAQQHLAEGRSRGGQTRKQQLGSEGYQEMGTKGGQTRKEQMG 256
Query: 54 TEGYQEMGRKGGLSTMEKSGGERAEEEGIDIDESKFK 90
EGYQEMGRKGGLSTM+KSG +RA EEGIDIDESKF+
Sbjct: 257 REGYQEMGRKGGLSTMDKSGEQRAAEEGIDIDESKFR 367
Score = 26.2 bits (56), Expect = 2.2
Identities = 24/83 (28%), Positives = 34/83 (40%), Gaps = 20/83 (24%)
Frame = +3
Query: 1 MASKQQNRQELEEK---------AKQGETV-----------VPGGTGGKSLEAQEHLAEG 40
MA +QQNR+ELEE+ Q E V V G G S Q+ +
Sbjct: 39 MAPQQQNREELEEREGLVKLLFLVAQVERVLKLNNTLLKEGVVEGRQGSSS*GQKDIKRW 218
Query: 41 RSKGGQTRKEQLGTEGYQEMGRK 63
K G+ K + G +G ++ K
Sbjct: 219 EQKEGRQGKSRWGEKGIKKWAVK 287
>TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalfa
(fragment), partial (91%)
Length = 800
Score = 30.4 bits (67), Expect = 0.11
Identities = 21/69 (30%), Positives = 32/69 (45%)
Frame = +2
Query: 2 ASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMG 61
A +Q L+E +Q G K L+ E+ E +G + +KEQL + QE+
Sbjct: 350 AMLRQQLHNLQESHRQIMGEELSGLTVKELQGLENQLEISLRGVRMKKEQLFMDEIQELN 529
Query: 62 RKGGLSTME 70
RKG + E
Sbjct: 530 RKGDIIHQE 556
>TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protein
{Corynebacterium glutamicum ATCC 13032}, partial (5%)
Length = 718
Score = 30.4 bits (67), Expect = 0.11
Identities = 16/39 (41%), Positives = 21/39 (53%)
Frame = +1
Query: 55 EGYQEMGRKGGLSTMEKSGGERAEEEGIDIDESKFKTGG 93
+G Q +GRK G K+GGE E G E+ +TGG
Sbjct: 226 KGNQGIGRKAGGEARHKAGGETC*EAGETYQETCQETGG 342
>BQ151355 homologue to GP|12620658|gb| ID556 {Bradyrhizobium japonicum},
partial (4%)
Length = 1420
Score = 28.9 bits (63), Expect = 0.33
Identities = 24/89 (26%), Positives = 33/89 (36%)
Frame = +1
Query: 9 QELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLST 68
Q ++ K+GET GG K+ + G Q KEQ G G + +
Sbjct: 208 QGQKKTKKEGETTEQGGERDKTKTTKPRTRPGGPPAAQAAKEQ-GRGGPAQPNNESRKPG 384
Query: 69 MEKSGGERAEEEGIDIDESKFKTGGGGGR 97
+ G + E E E K KT G R
Sbjct: 385 TPEDKGRKKEGERRQRTERKKKTRGSPAR 471
>TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19.70 -
Arabidopsis thaliana, partial (15%)
Length = 716
Score = 28.9 bits (63), Expect = 0.33
Identities = 23/73 (31%), Positives = 29/73 (39%)
Frame = -1
Query: 24 GGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEEEGID 83
GG+G +SL A AEG GG + L G + G GG ++ G
Sbjct: 512 GGSGSRSLAAGGSFAEGGGGGGCS----LAAGGSFDKGGGGGSCSLVAGG---------- 375
Query: 84 IDESKFKTGGGGG 96
F GGGGG
Sbjct: 374 ----SFDKGGGGG 348
>BF518891 weakly similar to GP|11993492|g ubiquitin-specific protease 26
{Arabidopsis thaliana}, partial (16%)
Length = 520
Score = 28.5 bits (62), Expect = 0.44
Identities = 17/46 (36%), Positives = 22/46 (46%), Gaps = 3/46 (6%)
Frame = -3
Query: 26 TGGKSLEAQEHLAEGRSKGGQTRKEQLGTE---GYQEMGRKGGLST 68
T SL EH E + GQTR+ L TE Y++ + G ST
Sbjct: 485 TSDNSLLHSEHSLESKGNVGQTRRSSLFTEVASSYKKSQKPSGTST 348
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 28.5 bits (62), Expect = 0.44
Identities = 23/74 (31%), Positives = 34/74 (45%)
Frame = +1
Query: 20 TVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEE 79
T V G G + + +E +GR +GG + GT+ E K S EK G+R EE
Sbjct: 973 TEV*GARGEREVGRRE---DGRERGGGVEAIRKGTQAGTETNEKAD-SRREKMRGKRREE 1140
Query: 80 EGIDIDESKFKTGG 93
+ + K+GG
Sbjct: 1141 R-----DGEAKSGG 1167
Score = 26.6 bits (57), Expect = 1.7
Identities = 26/104 (25%), Positives = 45/104 (43%), Gaps = 9/104 (8%)
Frame = +1
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
+A + ++ E K+G T +PG G K+ R + G+ E+ G++ +
Sbjct: 595 IARSVETEEQ*NETQKEGRTGMPGTRGRKNECVSAQYR*QRGR*GKRSGERGGSKTRKSR 774
Query: 61 GRK---GGLSTMEKS------GGERAEEEGIDIDESKFKTGGGG 95
R G + T E + G + E G E++ +TGGGG
Sbjct: 775 ARDRHIG*VETEESAR*IS*EGEQVRSERG---RETRSETGGGG 897
>TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog -
cucumber, partial (49%)
Length = 1486
Score = 27.7 bits (60), Expect = 0.74
Identities = 25/79 (31%), Positives = 36/79 (44%), Gaps = 1/79 (1%)
Frame = -2
Query: 8 RQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKG-GQTRKEQLGTEGYQEMGRKGGL 66
R+E EEKA+ GE + P + K ++ E G G T E + GR G +
Sbjct: 270 REEDEEKAR-GEAIAPARS*EKVADSVAKKLETSVIGPGLTALE---VKNASTEGRGGRV 103
Query: 67 STMEKSGGERAEEEGIDID 85
GGE + EEG+ +D
Sbjct: 102 MVGRGGGGEVSAEEGVVVD 46
>TC87120 similar to SP|P48724|IF5_PHAVU Eukaryotic translation initiation
factor 5 (eIF-5). [Kidney bean French bean] {Phaseolus
vulgaris}, partial (72%)
Length = 1974
Score = 27.7 bits (60), Expect = 0.74
Identities = 15/61 (24%), Positives = 27/61 (43%)
Frame = +1
Query: 27 GGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEEEGIDIDE 86
GGK +A + R K G+ E+L + + +KG + K+ + + G D D
Sbjct: 850 GGKDKKAMRRAEKERLKEGEAADEELKKLKKEAVKKKGAKESTAKTSSSKKKGNGSDEDH 1029
Query: 87 S 87
+
Sbjct: 1030T 1032
>TC92293 similar to SP|Q64733|FXB2_MOUSE Forkhead box protein B2
(Transcription factor FKH-4). [Mouse] {Mus musculus},
partial (4%)
Length = 626
Score = 27.3 bits (59), Expect = 0.97
Identities = 22/65 (33%), Positives = 30/65 (45%), Gaps = 5/65 (7%)
Frame = -1
Query: 40 GRSKGGQTRKEQLGTEGYQEMGRKGG---LSTMEKSGGERAEEEGIDIDESKFKTGG--G 94
GR +G T G +G +G GG ++ + ER EEE I K K+GG G
Sbjct: 536 GRERGRGTAD---GGDGIDSVGDDGGGDDVAMAYNNNREREEEEKGKIIRVKMKSGGCYG 366
Query: 95 GGRSQ 99
GG +
Sbjct: 365 GGEKR 351
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 27.3 bits (59), Expect = 0.97
Identities = 31/104 (29%), Positives = 39/104 (36%), Gaps = 12/104 (11%)
Frame = -1
Query: 6 QNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTE---------- 55
Q R+ EEK GE + G GG + R KG ++E+ G E
Sbjct: 563 QMRKGGEEKG--GE*MGTGARGGGGQAKERKRGRKRRKGENAKREKGGKERGKKERKKGA 390
Query: 56 --GYQEMGRKGGLSTMEKSGGERAEEEGIDIDESKFKTGGGGGR 97
G E KGG K +R G DE + GGG GR
Sbjct: 389 GNGIYEGEYKGGGGGGGKRKKKREGGRGKGKDEII*EKGGGMGR 258
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 27.3 bits (59), Expect = 0.97
Identities = 19/75 (25%), Positives = 30/75 (39%)
Frame = +1
Query: 23 PGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEEEGI 82
P GTGG+ + + G+ + Q + GT + G+K +K G + G
Sbjct: 430 PQGTGGEDTKTRP----GKQREPQQNPRERGTGHARTPGKKNTPPHTKKKGETHKKSPGG 597
Query: 83 DIDESKFKTGGGGGR 97
+ GGGG R
Sbjct: 598 RTPPPTPRPGGGGAR 642
>BQ143960 weakly similar to SP|P21997|SSGP_ Sulfated surface glycoprotein 185
(SSG 185). {Volvox carteri}, partial (13%)
Length = 1023
Score = 26.9 bits (58), Expect = 1.3
Identities = 16/35 (45%), Positives = 20/35 (56%), Gaps = 1/35 (2%)
Frame = -1
Query: 64 GGLSTMEKSGGE-RAEEEGIDIDESKFKTGGGGGR 97
GG E++GGE R E G +E K + GGGG R
Sbjct: 111 GGKRKWERNGGEMRGGEGGGGGEEGKGEGGGGGTR 7
>CA921832 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidopsis
thaliana}, partial (3%)
Length = 802
Score = 26.9 bits (58), Expect = 1.3
Identities = 21/83 (25%), Positives = 32/83 (38%), Gaps = 9/83 (10%)
Frame = -1
Query: 23 PGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGG---------LSTMEKSG 73
P GG ++ L R +G ++ E +E R G + + + G
Sbjct: 517 PDDDGGMEMQEPTRL---RDRGSGKKERDRERERERERDRLGRNKRRRNDRLMHGVREDG 347
Query: 74 GERAEEEGIDIDESKFKTGGGGG 96
GE EE I+ +E GGGG
Sbjct: 346 GEDTSEESINDEEDDDDEDGGGG 278
>TC82954 weakly similar to GP|8809591|dbj|BAA97142.1
emb|CAB82814.1~gene_id:MNB8.8~similar to unknown protein
{Arabidopsis thaliana}, partial (7%)
Length = 783
Score = 26.9 bits (58), Expect = 1.3
Identities = 14/38 (36%), Positives = 21/38 (54%)
Frame = +3
Query: 4 KQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGR 41
+++ R+ELEEKAK+ E E QEH ++ R
Sbjct: 555 EEKKRKELEEKAKKAEKDAEELRESSKREGQEHSSDLR 668
>BQ141694 similar to GP|12697907|db KIAA1681 protein {Homo sapiens}, partial
(2%)
Length = 1247
Score = 26.9 bits (58), Expect = 1.3
Identities = 17/46 (36%), Positives = 23/46 (49%), Gaps = 2/46 (4%)
Frame = -1
Query: 12 EEKAKQGETVVPGGTGGKSLEAQEHLAEGRS--KGGQTRKEQLGTE 55
E K K+ +T PG T AQE E R + GQ ++E+ G E
Sbjct: 146 ETKKKRDQTTGPGNTAPTGPPAQEEAEEVRKNRERGQKKREKGGGE 9
>BQ137257 similar to GP|21536912|gb| unknown {Arabidopsis thaliana}, partial
(15%)
Length = 1064
Score = 26.9 bits (58), Expect = 1.3
Identities = 14/58 (24%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Frame = +2
Query: 2 ASKQQNRQELEEKAKQGE----TVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTE 55
A ++++ + +++ K+GE PGGT G + + EG+ + G+ ++ G E
Sbjct: 89 ADQKRDENKKKKRKKEGE*EEKDQKPGGTKGGGGQKDKEAGEGKGERGRPKRRGEGRE 262
>BQ137240 similar to PIR|F75311|F753 ABC transporter ATP-binding protein -
Deinococcus radiodurans (strain R1), partial (4%)
Length = 1094
Score = 26.6 bits (57), Expect = 1.7
Identities = 15/39 (38%), Positives = 20/39 (50%)
Frame = +1
Query: 29 KSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLS 67
K E++E E K +TRKEQL G + R G +S
Sbjct: 715 KHKESREDEGEKNRKNTRTRKEQLNKTGRETATRVGRVS 831
>TC82317 similar to GP|2921209|gb|AAC04691.1| beta-ketoacyl-ACP synthase I
{Perilla frutescens}, partial (35%)
Length = 717
Score = 26.2 bits (56), Expect = 2.2
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = -1
Query: 64 GGLSTMEKSGGERAEEEGIDIDESKFKTGGGGGR 97
G L ++ GG+ ++G ES + +GGGGGR
Sbjct: 216 GILLSLRSGGGDGGRDDG----ESLWGSGGGGGR 127
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.299 0.126 0.331
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,632,002
Number of Sequences: 36976
Number of extensions: 16111
Number of successful extensions: 105
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 100
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 104
length of query: 101
length of database: 9,014,727
effective HSP length: 77
effective length of query: 24
effective length of database: 6,167,575
effective search space: 148021800
effective search space used: 148021800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (22.0 bits)
S2: 50 (23.9 bits)
Medicago: description of AC140721.4


