

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.3 + phase: 0 /pseudo
(1439 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
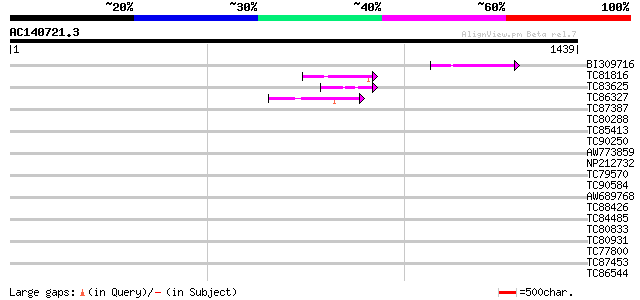
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 65 1e-10
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 53 1e-06
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 45 3e-04
TC86327 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 ... 44 3e-04
TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I... 42 0.002
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 42 0.002
TC85413 similar to PIR|S23737|S23737 proline-rich protein precur... 42 0.002
TC90250 similar to GP|20904925|gb|AAM30238.1 hypothetical protei... 41 0.003
AW773859 40 0.005
NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein 40 0.007
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 40 0.007
TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Ar... 39 0.011
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 39 0.011
TC88426 similar to GP|9758595|dbj|BAB09228.1 gene_id:MDF20.5~unk... 39 0.019
TC84485 homologue to GP|21624289|dbj|BAC01131. WfCYC2 {Wisteria ... 39 0.019
TC80833 weakly similar to PIR|D86175|D86175 hypothetical protein... 39 0.019
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 38 0.025
TC77800 similar to PIR|T02532|T02532 hypothetical protein At2g37... 38 0.032
TC87453 similar to GP|22093671|dbj|BAC06965. similar to bHLH (Ba... 38 0.032
TC86544 homologue to SP|P30706|PLSB_PEA Glycerol-3-phosphate acy... 37 0.042
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 65.5 bits (158), Expect = 1e-10
Identities = 72/227 (31%), Positives = 111/227 (48%), Gaps = 1/227 (0%)
Frame = +3
Query: 1068 KFASFKSLSMDLNRLVESGMRN*LLLFWLKTIYKLHLIIPCLSR-KLNHYSQFFWCMLMI 1126
++ SFK+L M +R V++G+ + L L + IY LI P L K+ H + MLM
Sbjct: 18 RYVSFKNLYMASSRQVDNGILSYLNLSFPLVIYNPLLIFPFLQNLKILHLLHC*Y-MLMT 194
Query: 1127 SFLLEIL*VNLIISSLFWILYSKSKT*VN*NTS*VLRLLIPNWGFLCVKGNIV*TYFLIQ 1186
F LE++ + + ++F ++ KSKT V + *VLRLL N F +K NI+ + I
Sbjct: 195 LF*LEMIYLKSNMLNVFLLIVLKSKTLVPYDIF*VLRLLEANKAFYLIKENILLNF*RIV 374
Query: 1187 EL*VPNQFLLHLISP*SCIMI*VLLLKMLLPTEDLWVDFFT*TLPDLISLSSLNNRVNSC 1246
+ + N LL +I *+ ++ M L ED + F * L LI NN N
Sbjct: 375 VILL*NPLLLLMIFL*NSTILIRPFTMMKLNIEDS*ANLFI*LLLVLIYPLLFNN*ANLF 554
Query: 1247 LNLHRLIIMLP*EF*STSKVVQEEAYFFHEILLSLYKDFQMLAGLAV 1293
NL + I L EF + K+ + Y ++L+ + Q+L G V
Sbjct: 555 RNLSKFTIRLLLEFYNI*KLPLPKDYSTQQLLI*NFLVLQILIGQLV 695
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 52.8 bits (125), Expect = 1e-06
Identities = 52/202 (25%), Positives = 93/202 (45%), Gaps = 12/202 (5%)
Frame = -2
Query: 743 FFIINSLILPFSKFL-----VVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIF 797
F ++ SL+ S+FL V + +L L+ + + Y + L++ +L +
Sbjct: 662 FSLLGSLVFILSRFLLLLFLVFFFLLLLFTFFRGFFYINIPYRYFTLFFLILLLLFLFLI 483
Query: 798 NLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLL 857
+ LLF+ +I +I+ L+L + +FL F L+ IL+ + + L L +LLL
Sbjct: 482 LI--------LLFLTSILTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLL 327
Query: 858 LLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFF------PI 911
+L + + L++ LLF +L L+ I FV + + +FLI F I
Sbjct: 326 ILLFITIFFFTLNILFLLFFFLLLFVLLILTLIYISFVLLLLILLFLFLIIFLY*FNLQI 147
Query: 912 IIFLTIILL-LLCLYIHNLSLN 932
+L +I + C +I NL N
Sbjct: 146 FFYLNLIFFYITC*FIINLDWN 81
Score = 37.7 bits (86), Expect = 0.032
Identities = 45/178 (25%), Positives = 78/178 (43%), Gaps = 4/178 (2%)
Frame = -2
Query: 358 LHLPAFPLQILLKVYLLLFLL*QVNLLFG*LIMVLMNIYVVILLHLVPFIE*NLFMFPYP 417
+++P + + LLLFL + LLF I+ + +++++L L+ F+ LF
Sbjct: 551 INIPYRYFTLFFLILLLLFLFLILILLFLTSILTFIIVFILVLFLLLVFLRFLLFTILVF 372
Query: 418 MVTLC*LILLVMSLSIPSFI*VMFFIHLILPSISSL*QNVVIH*LVSFIFLLKNVLYRTN 477
+ L+ L + L I FI + FF IL + F FLL VL
Sbjct: 371 VFFSLLLLFLFLLLLILLFITIFFFTLNIL--------------FLLFFFLLLFVL---- 246
Query: 478 NL*R*LVWLNRKMVYTSFIHHANLLILSFLLYLKVLVMLFLFPIVI----RIVNMYLT 531
L ++Y SF+ L+L LL+L +++ L+ F + I ++ Y+T
Sbjct: 245 --------LILTLIYISFV-----LLLLILLFLFLIIFLY*FNLQIFFYLNLIFFYIT 111
Score = 36.6 bits (83), Expect = 0.072
Identities = 32/115 (27%), Positives = 53/115 (45%), Gaps = 4/115 (3%)
Frame = -2
Query: 502 LILSFLLYLKVLVMLFLFP----IVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILIL 557
LIL L +L++LFL I++ I+ ++L + + L L+ F L L L
Sbjct: 515 LILLLLFLFLILILLFLTSILTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFL 336
Query: 558 LLIIIKMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IFGVLFQLLQFMVIDIF 612
LL+I+ + IF + L + +LL L F +L +L F+ + IF
Sbjct: 335 LLLILLFITIFFFTLNILFLLFFFLLLFVLLILTLIYISFVLLLLILLFLFLIIF 171
Score = 35.4 bits (80), Expect = 0.16
Identities = 32/92 (34%), Positives = 48/92 (51%), Gaps = 2/92 (2%)
Frame = -2
Query: 160 ISLIVLVFILVVVLHLKLLRFIEMRIRLCNF--LQD*MINFLLLELKFFSLILFLL*TKF 217
I ++VFILV+ L L LRF+ I + F L + LLL L F ++ F L F
Sbjct: 458 ILTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFITIFFFTLNILF 279
Query: 218 IL*LFRKKVIMLLLLL*VYLLIPPFKSMLLII 249
+L F ++L +LL + L+ F +LLI+
Sbjct: 278 LLFFF----LLLFVLLILTLIYISFVLLLLIL 195
Score = 32.3 bits (72), Expect = 1.4
Identities = 35/124 (28%), Positives = 64/124 (51%), Gaps = 7/124 (5%)
Frame = -2
Query: 298 MLLLKKILKMVQCLLLDWVLLQALLLLVFLKSN*LNLCP-CYNNQIWLFQHLLLHKLPPI 356
++L+ L + ++ ++L+ LLLVFL+ + + + + LF LLL L I
Sbjct: 488 LILILLFLTSILTFIIVFILV-LFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFI 312
Query: 357 TLHLPAFPLQILLKVYLLLFLL*QVNLL---FG*LIMVLMNIYVVILLH---LVPFIE*N 410
T+ + LL +LLLF+L + L+ F L+++L+ ++++I L+ L F N
Sbjct: 311 TIFFFTLNILFLLFFFLLLFVLLILTLIYISFVLLLLILLFLFLIIFLY*FNLQIFFYLN 132
Query: 411 LFMF 414
L F
Sbjct: 131 LIFF 120
Score = 32.3 bits (72), Expect = 1.4
Identities = 42/151 (27%), Positives = 73/151 (47%)
Frame = -2
Query: 89 TLFLSPLLKLLFFVILHLRFGLI*KNDSPKLIVFALQIYVLTSII*NKD*NLFWIISLK* 148
TLF LL L F+IL L F + +IVF L +++L ++ L+
Sbjct: 527 TLFFLILLLLFLFLILILLFLT---SILTFIIVFILVLFLL-------------LVFLR- 399
Query: 149 SPYGRNYLHIDISLIVLVFILVVVLHLKLLRFIEMRIRLCNFLQD*MINFLLLELKFFSL 208
+ +++V VF +++L L LL I + I + F + +L L FF L
Sbjct: 398 --------FLLFTILVFVFFSLLLLFLFLLLLILLFITIFFFTLN------ILFLLFFFL 261
Query: 209 ILFLL*TKFIL*LFRKKVIMLLLLL*VYLLI 239
+LF+L + ++ V++LL+LL ++L+I
Sbjct: 260 LLFVL--LILTLIYISFVLLLLILLFLFLII 174
Score = 31.6 bits (70), Expect = 2.3
Identities = 25/95 (26%), Positives = 44/95 (46%)
Frame = -2
Query: 155 YLHIDISLIVLVFILVVVLHLKLLRFIEMRIRLCNFLQD*MINFLLLELKFFSLILFLL* 214
Y++I L F+++++L L L+ + + F+ I F+L+ + FLL
Sbjct: 554 YINIPYRYFTLFFLILLLLFLFLILILLFLTSILTFI----IVFILVLFLLLVFLRFLLF 387
Query: 215 TKFIL*LFRKKVIMLLLLL*VYLLIPPFKSMLLII 249
T + F ++ L LLL + L I F L I+
Sbjct: 386 TILVFVFFSLLLLFLFLLLLILLFITIFFFTLNIL 282
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 44.7 bits (104), Expect = 3e-04
Identities = 40/148 (27%), Positives = 70/148 (47%), Gaps = 3/148 (2%)
Frame = -2
Query: 788 VIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPV 847
V + +S+ L +++F+ + L L+ LL +F+ F L L+ + L L +
Sbjct: 895 VSSISISLTDTLPSSFSSSIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLI 716
Query: 848 NLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLI 907
L + I L+ L+LL L LF LL L++++ ++ + + L IIFL
Sbjct: 715 -LLFITIFFFTLNILFLLFFFLLLFVLLI------LTLIYISFVLLLLILLFLFLIIFLY 557
Query: 908 FFPIIIFLTIILL---LLCLYIHNLSLN 932
F + IF + L+ + C +I NL N
Sbjct: 556 *FNLQIFFYLNLIFFYITC*FIINLDWN 473
Score = 37.7 bits (86), Expect = 0.032
Identities = 32/91 (35%), Positives = 51/91 (55%), Gaps = 1/91 (1%)
Frame = -2
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
+V ++L ++ RFL +L+F+ L L L LLL++ I + F IL LL
Sbjct: 835 IVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFITIFFFTLNILFLLFFF 656
Query: 847 VNLQSLMIL-LLLLHQLYLLLLLLHLFHLLF 876
+ L L+IL L+ + + LLL+LL LF ++F
Sbjct: 655 LLLFVLLILTLIYISFVLLLLILLFLFLIIF 563
Score = 35.0 bits (79), Expect = 0.21
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 2/88 (2%)
Frame = -2
Query: 164 VLVFILVVVLHLKLLRFIEMRIRLCNF--LQD*MINFLLLELKFFSLILFLL*TKFIL*L 221
++VFILV+ L L LRF+ I + F L + LLL L F ++ F L F+L
Sbjct: 838 IIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFITIFFFTLNILFLLFF 659
Query: 222 FRKKVIMLLLLL*VYLLIPPFKSMLLII 249
F ++L +LL + L+ F +LLI+
Sbjct: 658 F----LLLFVLLILTLIYISFVLLLLIL 587
Score = 31.6 bits (70), Expect = 2.3
Identities = 33/105 (31%), Positives = 54/105 (51%), Gaps = 7/105 (6%)
Frame = -2
Query: 317 LLQALLLLVFLKSN*LNLCP-CYNNQIWLFQHLLLHKLPPITLHLPAFPLQILLKVYLLL 375
+L LLLVFL+ + + + + LF LLL L IT+ + LL +LLL
Sbjct: 826 ILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFITIFFFTLNILFLLFFFLLL 647
Query: 376 FLL*QVNLL---FG*LIMVLMNIYVVILLH---LVPFIE*NLFMF 414
F+L + L+ F L+++L+ ++++I L+ L F NL F
Sbjct: 646 FVLLILTLIYISFVLLLLILLFLFLIIFLY*FNLQIFFYLNLIFF 512
Score = 31.6 bits (70), Expect = 2.3
Identities = 25/79 (31%), Positives = 46/79 (57%)
Frame = -2
Query: 161 SLIVLVFILVVVLHLKLLRFIEMRIRLCNFLQD*MINFLLLELKFFSLILFLL*TKFIL* 220
+++V VF +++L L LL I + I + F + +L L FF L+LF+L +
Sbjct: 778 TILVFVFFSLLLLFLFLLLLILLFITIFFFTLN------ILFLLFFFLLLFVL--LILTL 623
Query: 221 LFRKKVIMLLLLL*VYLLI 239
++ V++LL+LL ++L+I
Sbjct: 622 IYISFVLLLLILLFLFLII 566
>TC86327 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20
{Arabidopsis thaliana}, partial (50%)
Length = 3018
Score = 44.3 bits (103), Expect = 3e-04
Identities = 68/261 (26%), Positives = 119/261 (45%), Gaps = 16/261 (6%)
Frame = +3
Query: 656 LELIMALNSCYLLFMLHMAL-FTRNLV*KLPNKMVELKENINTF*MLVELCFISLN---- 710
L ++M L YL +L M L +R L + + +++L ++ L +L F+S
Sbjct: 354 LFVLMILFPAYLNMLLTMILKISR*L*IVMLHLLMKLASGMSVKRDLTKL-FLSTEPL*W 530
Query: 711 FLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSL-ILPFSKFLVVYVMLPLYILI 769
+LLP G++++ L+S H+ +I+ + ++ F V+ ++ L +L+
Sbjct: 531 WLLPMGVLIF*SLYS-------------HIPRLMLISPVELIKALLFTVLPQVVQLMLLM 671
Query: 770 EPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLL--FMKTISLTLILHPL---- 823
NC +L L S + L+ L + L FL +L F+K S T++L L
Sbjct: 672 L*NCFYQLV-LISILWMLMGNALWMLSLFLLLFLISSKVLKQFLKNFSQTVLLKDLWMIA 848
Query: 824 ---LLLIGNIFLIFPLYLILILLSLPVN-LQSLMILLLLLHQLYLLLLLLHLFHLLFPDI 879
LL + + PLY +L + P+ L SL I LL+LHQ + L HLF
Sbjct: 849 LFPCLLFHRVLVHLPLYHLLKMDLHPLLWLPSLQIQLLILHQKRKSIQLTHLFLT*KTAC 1028
Query: 880 LPETPLLHLTSKIIFVTIFKL 900
+P+ + S+ + V + L
Sbjct: 1029MPQMNSACIHSRFVLVLVHTL 1091
Score = 36.2 bits (82), Expect = 0.094
Identities = 29/118 (24%), Positives = 60/118 (50%)
Frame = +3
Query: 818 LILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFP 877
+++ P+ L+ +F + P + L+LL L L+++ +L + L +L LF LLF
Sbjct: 591 MLISPVELIKALLFTVLPQVVQLMLLML*NCFYQLVLISILWMLMGNALWMLSLFLLLF- 767
Query: 878 DILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLYIHNLSLNPML 935
L + +L K T+ L+ + + FP ++F +++ L ++ + L+P+L
Sbjct: 768 --LISSKVLKQFLKNFSQTVL-LKDLWMIALFPCLLFHRVLVHLPLYHLLKMDLHPLL 932
>TC87387 homologue to PIR|T47775|T47775 hypothetical protein F24I3.230 -
Arabidopsis thaliana, partial (14%)
Length = 1074
Score = 42.0 bits (97), Expect = 0.002
Identities = 50/183 (27%), Positives = 85/183 (46%), Gaps = 6/183 (3%)
Frame = -2
Query: 756 FLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTIS 815
FL ++V L L +L+ + ++ L + + S++ + ++F L R + F + +
Sbjct: 716 FLGIFVFLFLVLLLT-SISIALIISVTDIGSIIFGFFVFLLFLLLRLVHFRRTNFTRFLV 540
Query: 816 LTLILHPLLLLIGNIFLIFPLYLILILLSLPV---NLQSLMILLLLLHQLYLLLLLLHLF 872
L +L LL IF +F + L L+L +L + + L ILLL L +L++ LH
Sbjct: 539 LLFLLIRLLHFRRTIFTLFLVLLFLLLFNLLLIRCSSSILFILLLFLLLFFLVIFNLHSR 360
Query: 873 HLLFPDILPETPLLHLTSKIIFVT---IFKLRHIIFLIFFPIIIFLTIILLLLCLYIHNL 929
+ F LH I F + + R I +F + F I+L LL L+I NL
Sbjct: 359 VIYFLG-------LHFFFAINFSNFGLLSRSRAIGRFMFLSLTPFSIILLNLLTLFISNL 201
Query: 930 SLN 932
L+
Sbjct: 200 FLH 192
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 42.0 bits (97), Expect = 0.002
Identities = 53/225 (23%), Positives = 98/225 (43%), Gaps = 38/225 (16%)
Frame = -1
Query: 773 CNLELESLYS*VTSLVIRVLLSM------------IFNLERFLCQDMLLFMKT----ISL 816
CNL++ ++ + VT IR++ + +F + F M + + ISL
Sbjct: 995 CNLQIRNILTPVT--FIRMICFVFRFSGIL*A*IRVFQITCFSASFMSIMCHSTRGIISL 822
Query: 817 TLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQL---------YLLLL 867
+ ++L+ +F PL+LIL++ L + + + L +H L +L+++
Sbjct: 821 VMFFPSFMILV--VFN*TPLFLILLVFILRLIITLFFFVFLFIHNLILTIIICL*FLMII 648
Query: 868 LLHL------FHLL-FPDILPETPLLHLTSKIIFVTIF---KLRHIIFLIFFPIII---F 914
++L FHL F I LLH S +IF I HI+F FF ++ F
Sbjct: 647 FIYLNISSFIFHLFTFAFIF----LLHFCSMLIFFFILFFSPFMHIVFTFFFSFVLVSFF 480
Query: 915 LTIILLLLCLYIHNLSLNPMLKQVNMSVGCMLCKLNFKLLRKLVL 959
++L ++ +I ++ N + + ++C F L R VL
Sbjct: 479 SFLLLFIVFTFILHIVYNMFIFLFFLMPFILIC---FNLTRMKVL 354
>TC85413 similar to PIR|S23737|S23737 proline-rich protein precursor -
kidney bean, partial (56%)
Length = 1139
Score = 42.0 bits (97), Expect = 0.002
Identities = 40/124 (32%), Positives = 61/124 (48%), Gaps = 14/124 (11%)
Frame = +2
Query: 811 MKTISLTLILH-PLLLLIGNIFLIF-PLY-----LILILLSLPVNLQSLMILLLLLHQLY 863
+ TI+ T H PLLLL+ + L+F PLY L+L LL+ + + +IL LLL L
Sbjct: 185 LTTITTTTFTHQPLLLLLLLLLLLFTPLYTLLTILLLYLLNHQLTATTTIILTLLLLHLS 364
Query: 864 LLLLLLHLFH-------LLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLT 916
LLL H+ H LLF L T LL T I + + + ++F + +
Sbjct: 365 THLLLQHIHHYTHPFTPLLFQLTLHYTHLLQHTHHCILLPSLEAL*LFKVLFMSNLASML 544
Query: 917 IILL 920
+++L
Sbjct: 545 VLIL 556
>TC90250 similar to GP|20904925|gb|AAM30238.1 hypothetical protein
{Methanosarcina mazei Goe1}, partial (13%)
Length = 740
Score = 41.2 bits (95), Expect = 0.003
Identities = 41/139 (29%), Positives = 70/139 (49%), Gaps = 7/139 (5%)
Frame = +2
Query: 791 VLLSMIFNLERFLCQDMLLFMKT---ISLTLILHPLLLLIGNIFLIFPLYLILILLSLPV 847
+LLS+ L L + L F T I L L+ H LL + N+ + + L+ IL L +
Sbjct: 122 LLLSLSPLLVHSLLRTALKFTLTNLLILLNLLTHLNLLTLLNLLTLLNILLLNILTLLNI 301
Query: 848 NLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHII--- 904
L +++ LL +L LL +L L ++L +IL +L L + + + I L +I+
Sbjct: 302 LLLNILTLLNIL-----LLNILTLLNILLLNILTLLNILTLPNILTILNILTLPNILTIL 466
Query: 905 -FLIFFPIIIFLTIILLLL 922
L+ PI+I +IL+ +
Sbjct: 467 NILMLLPILILT*VILICI 523
>AW773859
Length = 538
Score = 40.4 bits (93), Expect = 0.005
Identities = 50/147 (34%), Positives = 68/147 (46%), Gaps = 6/147 (4%)
Frame = -1
Query: 470 KNVLYRTNNL*R*LVWLNRKMVYTSFIHHANLL----ILSFLLYLKVLVMLFLFPIVIRI 525
K+ +R *R*LV +N S H + L +L F L L +L
Sbjct: 391 KSY*FRKRGA*R*LVRVNYLKDCISLPHKIHPLPP*HLLKFNLNLNLL------------ 248
Query: 526 VNMYLTMPYGTLDLVICLIKDFI*CLACILILLLII-IKML-VIFAILPSTNIYLSILVP 583
YL P+GTLD I I++ CLA I+I LL + IK+L V + I+P YL LV
Sbjct: 247 --FYLKKPFGTLDWDIYPIEN---CLAYIVIFLL*L*IKILFVTYVIIPDIKSYLFSLVQ 83
Query: 584 HMLLLNLNFSI*IFGVLFQLLQFMVID 610
L +N I+GV F F++ D
Sbjct: 82 IELPNVMNCFTLIYGVPFPPNLFIIKD 2
>NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein
Length = 576
Score = 40.0 bits (92), Expect = 0.007
Identities = 42/133 (31%), Positives = 68/133 (50%), Gaps = 13/133 (9%)
Frame = -3
Query: 807 MLLFMKTISLTLILHPLLLLIGNI--FLIFPLYLILILLSLPVNLQSLMILLLLLHQLYL 864
+LLF + LTL+L L L ++ + L L+++LL L V L L++ L+LL L L
Sbjct: 502 LLLFWQLCFLTLLLEQLTFLFLHLE*VRLLLLSLVMVLLLLVVVLLVLVLFLVLL--LVL 329
Query: 865 LLLLLHLFHLLFPDILP-ETPL--LHLTSKIIFVTIFKLR-------HIIF-LIFFPIII 913
LL+LL L L FP +L P LHL + L H+++ + P ++
Sbjct: 328 LLVLLVLVELQFP*LLL*GHPFL*LHLVLHLFL*LHLVLHPFL*LHLHLVYPFL*HPFLL 149
Query: 914 FLTIILLLLCLYI 926
+ ++LL+L L +
Sbjct: 148 VVVVVLLVLVLLV 110
Score = 30.4 bits (67), Expect = 5.2
Identities = 20/73 (27%), Positives = 39/73 (53%), Gaps = 1/73 (1%)
Frame = -3
Query: 855 LLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLT-SKIIFVTIFKLRHIIFLIFFPIII 913
LLLL QL L LLL LF LHL +++ +++ + ++ ++ +++
Sbjct: 505 LLLLFWQLCFLTLLLEQLTFLF---------LHLE*VRLLLLSLVMVLLLLVVVLLVLVL 353
Query: 914 FLTIILLLLCLYI 926
FL ++L+LL + +
Sbjct: 352 FLVLLLVLLLVLL 314
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 40.0 bits (92), Expect = 0.007
Identities = 39/112 (34%), Positives = 57/112 (50%), Gaps = 11/112 (9%)
Frame = -3
Query: 824 LLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPET 883
LL+I N+ L+ L L+L+L+ ++LLLLL L LLLLLLH+ L+ +L
Sbjct: 685 LLMIYNLLLLL*LLLLLLLM---------LLLLLLLLLLLLLLLLLHMIILISE*VLMLK 533
Query: 884 PLLH-----------LTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCL 924
LL LT I++ ++ +LR + LI I + IL LL L
Sbjct: 532 KLLELLCLGLRLRWMLTFLILWKSLMRLRLLQHLILH*SSITIGEILRLLKL 377
Score = 33.1 bits (74), Expect = 0.80
Identities = 32/85 (37%), Positives = 44/85 (51%), Gaps = 6/85 (7%)
Frame = -3
Query: 793 LSMIFNLERF--LCQDMLLFMKTISLTLILHPLLLLIGNIFLI----FPLYLILILLSLP 846
L MI+NL L +LL + + L L+L LLLL+ I LI L +L LL L
Sbjct: 685 LLMIYNLLLLL*LLLLLLLMLLLLLLLLLLLLLLLLLHMIILISE*VLMLKKLLELLCLG 506
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHL 871
+ L+ ++ L+L L L LL HL
Sbjct: 505 LRLRWMLTFLILWKSLMRLRLLQHL 431
Score = 30.4 bits (67), Expect = 5.2
Identities = 27/95 (28%), Positives = 53/95 (55%), Gaps = 3/95 (3%)
Frame = -3
Query: 852 LMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLR---HIIFLIF 908
L++LLLL+ L LLLLLL L LL I+ + + + K++ + LR + FLI
Sbjct: 649 LLLLLLLMLLLLLLLLLLLLLLLLLHMIILISE*VLMLKKLLELLCLGLRLRWMLTFLIL 470
Query: 909 FPIIIFLTIILLLLCLYIHNLSLNPMLKQVNMSVG 943
+ ++ L ++ L+ L+ ++++ +L+ + + G
Sbjct: 469 WKSLMRLRLLQHLI-LH*SSITIGEILRLLKLKQG 368
>TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Arabidopsis
thaliana}, partial (18%)
Length = 686
Score = 39.3 bits (90), Expect = 0.011
Identities = 28/97 (28%), Positives = 53/97 (53%), Gaps = 4/97 (4%)
Frame = +1
Query: 853 MILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFP-- 910
++L LLL L L LLLHL LL P + P+ PL ++++ ++ + L+
Sbjct: 103 LLLRLLLPPLLHLPLLLHLPQLLPPKV-PQIPLPIPEAELVAAVAVMMKKTVMLLIVVIG 279
Query: 911 -IIIFLTIILLLLCL-YIHNLSLNPMLKQVNMSVGCM 945
I+I + I+++ +CL I + + P+LK + + + C+
Sbjct: 280 LILIMMMIMMIEICLVMIMKIIVKPLLKVLTLFLYCL 390
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 39.3 bits (90), Expect = 0.011
Identities = 37/113 (32%), Positives = 51/113 (44%)
Frame = +2
Query: 943 GCMLCKLNFKLLRKLVLGNLWIYHQMLSLLDVDGFTKSNFMLMALLRDIKQDWLPKVTIK 1002
GC L KL+ KL G L+++ LL GF + M LL ++K D +V++K
Sbjct: 98 GCKL*KLSTKL*LITKHGILFLFLHTRKLLAASGFIELKKTQMDLLTNLKPD*WLRVSVK 277
Query: 1003 LRAWIILIHILQLLNSQQSDL*LLYLPFIICIYINLMSIMPFFMVNFRKMCTC 1055
I L L N S L I + L+S MPF MV F++ C C
Sbjct: 278 HLDVITLKLSLL**NLSLSGSFSPLLSLINGKFNKLISTMPF*MVFFKRKCIC 436
>TC88426 similar to GP|9758595|dbj|BAB09228.1 gene_id:MDF20.5~unknown
protein {Arabidopsis thaliana}, partial (56%)
Length = 1295
Score = 38.5 bits (88), Expect = 0.019
Identities = 31/83 (37%), Positives = 46/83 (55%)
Frame = +1
Query: 838 LILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTI 897
+IL+ LP+ L L++LLLLL L LLL+ L L L F L L KI+F+
Sbjct: 184 IILLFNLLPLFLLPLLLLLLLLQLLLLLLVPLLLQGLSFGYQLHIVMLAPQYLKILFLLY 363
Query: 898 FKLRHIIFLIFFPIIIFLTIILL 920
L I+FL++ ++ TI++L
Sbjct: 364 KVLLKILFLLYKVLLKGSTIMML 432
Score = 30.4 bits (67), Expect = 5.2
Identities = 24/87 (27%), Positives = 44/87 (49%), Gaps = 7/87 (8%)
Frame = +1
Query: 792 LLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIG-------NIFLIFPLYLILILLS 844
++ ++FNL +LL + + L L+L LLL G +I ++ P YL ++ L
Sbjct: 181 IIILLFNLLPLFLLPLLLLLLLLQLLLLLLVPLLLQGLSFGYQLHIVMLAPQYLKILFLL 360
Query: 845 LPVNLQSLMILLLLLHQLYLLLLLLHL 871
V L+ L +L +L + +++L L
Sbjct: 361 YKVLLKILFLLYKVLLKGSTIMMLKDL 441
>TC84485 homologue to GP|21624289|dbj|BAC01131. WfCYC2 {Wisteria
floribunda}, partial (45%)
Length = 728
Score = 38.5 bits (88), Expect = 0.019
Identities = 31/106 (29%), Positives = 53/106 (49%)
Frame = +1
Query: 853 MILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPII 912
+ L LH L LLLLLLH F L + +L + + +T F + H FL + I
Sbjct: 31 LTLHFFLHHLLLLLLLLHSFLFL-------S*ILKMLLQTTTITHFFMIHFQFLT-YQFI 186
Query: 913 IFLTIILLLLCLYIHNLSLNPMLKQVNMSVGCMLCKLNFKLLRKLV 958
I L +I + L ++ + +LK++ + V ++ L F+ L K++
Sbjct: 187 IILILIFITLQIFQKP*PIWLLLKKITIIVTTIIITLLFQCLNKIL 324
>TC80833 weakly similar to PIR|D86175|D86175 hypothetical protein [imported]
- Arabidopsis thaliana, partial (60%)
Length = 709
Score = 38.5 bits (88), Expect = 0.019
Identities = 33/89 (37%), Positives = 50/89 (56%), Gaps = 6/89 (6%)
Frame = +2
Query: 820 LHPLLLLIGNIFLIFPLYLILIL-LSLPVNLQSLMILLLLLHQL---YLLLLLLHLFHLL 875
L L++L+ IF++ L+ I ++ + L ++L + LLL LH L +LLLLLL LL
Sbjct: 287 LQELMMLLLFIFVVIELFSISLMNIKLLIHLLHYLCLLLCLHHLLLIHLLLLLLLPLFLL 466
Query: 876 FPDILPETPLLHLTSKII--FVTIFKLRH 902
P ++ L+ L S II F FK+ H
Sbjct: 467 TPLMM---VLMSLNSWIISCFKNFFKMEH 544
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 38.1 bits (87), Expect = 0.025
Identities = 34/80 (42%), Positives = 44/80 (54%)
Frame = -2
Query: 799 LERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLL 858
L + L +L K + L L L LLLL L+ L+L L +L +NL L+I LL
Sbjct: 778 LRKLLMLQLL*LRKLLLLLLQLRKLLLLKLRELLL----LLLELWNLLLNLGKLLI-LLQ 614
Query: 859 LHQLYLLLLLLHLFHLLFPD 878
L +L LLLLLLHL L+ D
Sbjct: 613 LWKLLLLLLLLHLLLQLWKD 554
Score = 32.7 bits (73), Expect = 1.0
Identities = 31/73 (42%), Positives = 41/73 (55%), Gaps = 4/73 (5%)
Frame = -2
Query: 807 MLLFMKTISLTLI----LHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQL 862
+LL K + L L+ L LLL + + L+ L+L+LL L L +L LL+LL
Sbjct: 787 LLLLRKLLMLQLL*LRKLLLLLLQLRKLLLLKLRELLLLLLELWNLLLNLGKLLILLQLW 608
Query: 863 YLLLLLLHLFHLL 875
LLLLLL L HLL
Sbjct: 607 KLLLLLL-LLHLL 572
>TC77800 similar to PIR|T02532|T02532 hypothetical protein At2g37660
[imported] - Arabidopsis thaliana, partial (73%)
Length = 1379
Score = 37.7 bits (86), Expect = 0.032
Identities = 29/113 (25%), Positives = 60/113 (52%), Gaps = 2/113 (1%)
Frame = +2
Query: 803 LCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNL-QSLMILLLLLHQ 861
L + L + T+ L +L L L+ ++FL+F L + ++ ++ Q +I L LLH
Sbjct: 119 LTSKLPLIITTMLLCELL*C*LWLLLHVFLLFLLPHFFLTNAINIHWWQGQLIYLFLLHH 298
Query: 862 L-YLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIII 913
+LL++LLH FHL + +L E + + ++ + L+ ++ L+ +++
Sbjct: 299 *GFLLVILLHWFHLHYQGVLRE---VEIEEVLLLLLWLSLQRVLCLLLVLVVV 448
>TC87453 similar to GP|22093671|dbj|BAC06965. similar to bHLH
(Basic-helix-loop-helix) transcription factor, partial
(26%)
Length = 1879
Score = 37.7 bits (86), Expect = 0.032
Identities = 46/154 (29%), Positives = 68/154 (43%), Gaps = 10/154 (6%)
Frame = +1
Query: 816 LTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
L I L+ +GNI L+ L+ ILIL S + L++ ++ Q Y +L L L
Sbjct: 535 LVTI*RNLIQQVGNIILVVQLHSILILRST*MGTVKLLLRIMKG*QNYPIL*ALGL---- 702
Query: 876 FPDILPETPLLHLTSKIIFVTIFKLRHIIFLI----FFPIIIFLTIILLLLCLYIHNLSL 931
L HLT K++ I K I LI PI I +T ++L + I N L
Sbjct: 703 ---------LPHLTLKLVATLIHKQITICLLI*IILIRPIWIIITTHMILTLIVISNYLL 855
Query: 932 ---NPMLKQVNMSVGCMLCKLNFKL---LRKLVL 959
+ +L + + C L L FK LRK ++
Sbjct: 856 VIPHHVLLEKE*VIKCSLVALIFKT*IRLRKNII 957
>TC86544 homologue to SP|P30706|PLSB_PEA Glycerol-3-phosphate
acyltransferase chloroplast precursor (EC 2.3.1.15)
(GPAT). [Garden pea], partial (85%)
Length = 1951
Score = 37.4 bits (85), Expect = 0.042
Identities = 30/83 (36%), Positives = 46/83 (55%), Gaps = 6/83 (7%)
Frame = +3
Query: 798 NLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILL- 856
NL++ L LL SL L H LLLL+ + L+ L+++L L + +Q L++LL
Sbjct: 63 NLQQCLQHHFLLLQPHFSLHLKPHILLLLLLLLLLLRYLFVVLSLFIIFDLMQQLLLLL* 242
Query: 857 -----LLLHQLYLLLLLLHLFHL 874
LLLH + LLL +L + +L
Sbjct: 243 QLLELLLLHIVLLLLSILIIKNL 311
Score = 32.7 bits (73), Expect = 1.0
Identities = 29/104 (27%), Positives = 59/104 (55%), Gaps = 2/104 (1%)
Frame = +3
Query: 766 YILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLL 825
++L++P+ +L L+ I +LL ++ L R+L + LF+ + ++ LLL
Sbjct: 90 FLLLQPHFSLHLKPH--------ILLLLLLLLLLLRYLFVVLSLFI----IFDLMQQLLL 233
Query: 826 LIGNIFLIFPLYLILILLSLPV--NLQSLMILLLLLHQLYLLLL 867
L+ + + L+++L+LLS+ + NL+ +++ LL L+LL L
Sbjct: 234 LL*QLLELLLLHIVLLLLSILIIKNLKKFLLIWRLL--LFLLAL 359
Score = 30.0 bits (66), Expect = 6.8
Identities = 35/112 (31%), Positives = 55/112 (48%), Gaps = 12/112 (10%)
Frame = +3
Query: 346 QHLLLHKLPPITLHLPAFPLQILLKVYLLL------------FLL*QVNLLFG*LIMVLM 393
QH L P +LHL L +LL + LLL F L Q LL * ++ L+
Sbjct: 81 QHHFLLLQPHFSLHLKPHILLLLLLLLLLLRYLFVVLSLFIIFDLMQQLLLLL*QLLELL 260
Query: 394 NIYVVILLHLVPFIE*NLFMFPYPMVTLC*LILLVMSLSIPSFI*VMFFIHL 445
+++V+LL + I+ NL F + + L+L +++LS + MFF+ L
Sbjct: 261 LLHIVLLLLSILIIK-NLKKF----LLIWRLLLFLLALSSMPEMNKMFFLEL 401
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.356 0.163 0.531
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,815,308
Number of Sequences: 36976
Number of extensions: 1077939
Number of successful extensions: 23113
Number of sequences better than 10.0: 247
Number of HSP's better than 10.0 without gapping: 7751
Number of HSP's successfully gapped in prelim test: 1677
Number of HSP's that attempted gapping in prelim test: 11185
Number of HSP's gapped (non-prelim): 13772
length of query: 1439
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1331
effective length of database: 5,021,319
effective search space: 6683375589
effective search space used: 6683375589
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC140721.3


