

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.11 + phase: 0
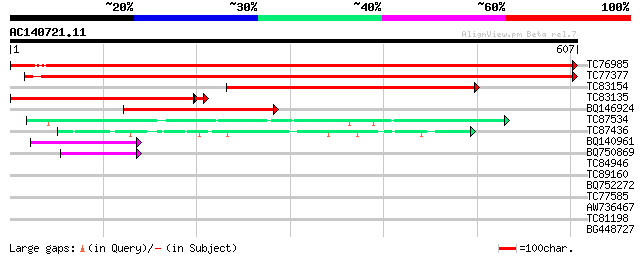
(607 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76985 homologue to GP|17225598|gb|AAL37492.1 pyruvate decarbox... 1067 0.0
TC77377 homologue to SP|P51850|DCP1_PEA Pyruvate decarboxylase i... 953 0.0
TC83154 similar to SP|P51851|DCP2_PEA Pyruvate decarboxylase iso... 539 e-153
TC83135 similar to GP|10187157|emb|CAC09052. cDNA~Strawberry pyr... 401 e-115
BQ146924 similar to GP|10732644|gb pyruvate decarboxylase 1 {Vit... 275 4e-74
TC87534 similar to SP|P17597|ILVB_ARATH Acetolactate synthase c... 85 7e-17
TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyas... 62 6e-10
BQ140961 similar to SP|P09342|ILV1 Acetolactate synthase I chlo... 60 2e-09
BQ750869 weakly similar to GP|21205303|db alpha-acetolactate syn... 50 2e-06
TC84946 similar to GP|16245|emb|CAA35887.1|| precursor acetolact... 40 0.002
TC89160 similar to GP|9759326|dbj|BAB09835.1 contains similarity... 32 0.92
BQ752272 GP|1742479|dbj ORF_ID:o302#2~similar to {SwissProt Acce... 31 1.2
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 30 3.5
AW736467 similar to PIR|T05689|T0 hypothetical protein F20M13.17... 29 6.0
TC81198 similar to GP|21553392|gb|AAM62485.1 unknown {Arabidopsi... 28 7.8
BG448727 similar to PIR|T08924|T08 hypothetical protein T15N24.3... 28 7.8
>TC76985 homologue to GP|17225598|gb|AAL37492.1 pyruvate decarboxylase
{Fragaria x ananassa}, partial (94%)
Length = 2130
Score = 1067 bits (2760), Expect = 0.0
Identities = 521/607 (85%), Positives = 562/607 (91%)
Frame = +1
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGI 60
M+T LGSL+ K ND+++ N T +TIQ PST ++A+S++TLG HLARRLV+VG+
Sbjct: 121 METMLGSLDIAKPTSNDVVSCKQQNST-ATIQ--PST-AIATSDATLGRHLARRLVQVGV 288
Query: 61 TDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGL 120
TD+F+VPGDFNLTLLDHLI EP+L IGCCNELNAGYAADGYARSRGVGACVVTFTVGGL
Sbjct: 289 TDVFSVPGDFNLTLLDHLINEPELNLIGCCNELNAGYAADGYARSRGVGACVVTFTVGGL 468
Query: 121 SVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVV 180
SV+NAIAGAYSENLP+ICIVGGPNSND+GT+RILHHTIG+PDFSQELRCFQTVTC+QAVV
Sbjct: 469 SVLNAIAGAYSENLPLICIVGGPNSNDYGTSRILHHTIGVPDFSQELRCFQTVTCFQAVV 648
Query: 181 NNLEDAHEMIDTAISTALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMGLE 240
NNLEDAHE+IDTAISTALKESKPVYISI CNL IPHPTFSR+PVPFSL+PKLSN MGLE
Sbjct: 649 NNLEDAHELIDTAISTALKESKPVYISIGCNLPGIPHPTFSRDPVPFSLAPKLSNHMGLE 828
Query: 241 AAVEAAAEVLNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDHKH 300
AAVEAAAE LNKAVKPVLV GPKLRVAKA +AF+ELAD S Y +VMPSAKG++PE H H
Sbjct: 829 AAVEAAAEFLNKAVKPVLVGGPKLRVAKASDAFVELADASGYALAVMPSAKGMVPEHHPH 1008
Query: 301 FIGTFWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIGN 360
FIGT+WGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVV+ N
Sbjct: 1009FIGTYWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVVAN 1188
Query: 361 GPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHIQN 420
GP FGC+LMKDFL ALAKRLK NN AYENY RIFVP+G P+KS P+EPLRVNV+FQHIQ
Sbjct: 1189GPAFGCILMKDFLKALAKRLKHNNAAYENYHRIFVPDGKPLKSAPKEPLRVNVMFQHIQQ 1368
Query: 421 MLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIA 480
MLS ETAVIAETGDSWFNCQKLKLP+GCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIA
Sbjct: 1369MLSRETAVIAETGDSWFNCQKLKLPEGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIA 1548
Query: 481 CIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAI 540
CIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGL+DAI
Sbjct: 1549CIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLIDAI 1728
Query: 541 HNGQGHCWTTKVFCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSSAN 600
HNG+G CWTTKVFCEEELVEAI TATGPKKD LCFIEVIVHKDDTSKELLEWGSRVS+AN
Sbjct: 1729HNGEGKCWTTKVFCEEELVEAIATATGPKKDSLCFIEVIVHKDDTSKELLEWGSRVSAAN 1908
Query: 601 SRPPNPQ 607
SRPPNPQ
Sbjct: 1909SRPPNPQ 1929
>TC77377 homologue to SP|P51850|DCP1_PEA Pyruvate decarboxylase isozyme 1
(EC 4.1.1.1) (PDC). [Garden pea] {Pisum sativum},
partial (95%)
Length = 2128
Score = 953 bits (2463), Expect = 0.0
Identities = 456/592 (77%), Positives = 521/592 (87%)
Frame = +2
Query: 16 NDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGITDIFTVPGDFNLTLL 75
N + T+P S+ +PST + + T+G HLARRLVE+G+ D+F+VPGDFNLTLL
Sbjct: 146 NSMETSPPSS--------APSTVRPFTCDGTMGGHLARRLVEIGVRDVFSVPGDFNLTLL 301
Query: 76 DHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGLSVINAIAGAYSENLP 135
DHLIAEP+L +GCCNELNAGYAADGYAR++GVGACVVTFTVGGLS++NAIAGAYSENLP
Sbjct: 302 DHLIAEPELNLVGCCNELNAGYAADGYARAKGVGACVVTFTVGGLSILNAIAGAYSENLP 481
Query: 136 VICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNNLEDAHEMIDTAIS 195
VICIVGGPNSND+GTNRILHHTIGLPDFSQELRCFQT+TC+QAVVNNLEDAHE+IDTAIS
Sbjct: 482 VICIVGGPNSNDYGTNRILHHTIGLPDFSQELRCFQTITCFQAVVNNLEDAHELIDTAIS 661
Query: 196 TALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMGLEAAVEAAAEVLNKAVK 255
TALKESKPVYISI CNL +IPHPTF+R+PVPF L+P++SN GLEAAVE AA LNKAVK
Sbjct: 662 TALKESKPVYISIGCNLPAIPHPTFARDPVPFFLAPRVSNQEGLEAAVEEAAAFLNKAVK 841
Query: 256 PVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVSTAFCA 315
PV+V GPKLRVAKA +AF+E A+ S YP +VMPS KGL+PE+H HFIGT+WGAVST++C
Sbjct: 842 PVIVGGPKLRVAKAQKAFMEFAEASGYPIAVMPSGKGLVPENHPHFIGTYWGAVSTSYCG 1021
Query: 316 EIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIGNGPTFGCVLMKDFLSA 375
EIVESADAY+F GPIFNDYSSVGYSLL+KKEK++IVQP+RV IGNG + G V M DFL+A
Sbjct: 1022EIVESADAYVFVGPIFNDYSSVGYSLLIKKEKSLIVQPNRVTIGNGLSLGWVFMADFLTA 1201
Query: 376 LAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHIQNMLSSETAVIAETGDS 435
L+K++K+N A ENY RI+VP G+P+K E EPLRVNVLF+HIQ +LS +TAVIAETGDS
Sbjct: 1202LSKKVKKNTAAVENYRRIYVPPGIPLKWEKDEPLRVNVLFKHIQELLSGDTAVIAETGDS 1381
Query: 436 WFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVS 495
WFNCQKL+LP+ CGYEFQMQYGSIGWSVGATLGYAQA KRVIACIGDGSFQVTAQD+S
Sbjct: 1382WFNCQKLRLPENCGYEFQMQYGSIGWSVGATLGYAQAATNKRVIACIGDGSFQVTAQDIS 1561
Query: 496 TMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAIHNGQGHCWTTKVFCE 555
TM+RCGQK+IIFLINNGGYTIEVEIHDGPYNVIKNW+YT V AIHNGQG CWT KV E
Sbjct: 1562TMIRCGQKSIIFLINNGGYTIEVEIHDGPYNVIKNWDYTRFVSAIHNGQGKCWTAKVRTE 1741
Query: 556 EELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSSANSRPPNPQ 607
E+L+EAI TATG +KD LCFIEV HKDDTSKELLEWGSRV++ANSRPPNPQ
Sbjct: 1742EDLIEAIATATGTEKDSLCFIEVFAHKDDTSKELLEWGSRVAAANSRPPNPQ 1897
>TC83154 similar to SP|P51851|DCP2_PEA Pyruvate decarboxylase isozyme 2 (EC
4.1.1.1) (PDC) (Fragment). [Garden pea] {Pisum sativum},
partial (68%)
Length = 827
Score = 539 bits (1388), Expect = e-153
Identities = 269/271 (99%), Positives = 269/271 (99%)
Frame = +1
Query: 233 LSNPMGLEAAVEAAAEVLNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKG 292
LSNPMGLEAAVEAAAE LNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKG
Sbjct: 1 LSNPMGLEAAVEAAAEFLNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKG 180
Query: 293 LIPEDHKHFIGTFWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQ 352
LIPEDHKHFIGTFWGAVSTAF AEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQ
Sbjct: 181 LIPEDHKHFIGTFWGAVSTAFGAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQ 360
Query: 353 PDRVVIGNGPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVN 412
PDRVVIGNGPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVN
Sbjct: 361 PDRVVIGNGPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVN 540
Query: 413 VLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQA 472
VLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQA
Sbjct: 541 VLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQA 720
Query: 473 VPEKRVIACIGDGSFQVTAQDVSTMLRCGQK 503
VPEKRVIACIGDGSFQVTAQDVSTMLRCGQK
Sbjct: 721 VPEKRVIACIGDGSFQVTAQDVSTMLRCGQK 813
>TC83135 similar to GP|10187157|emb|CAC09052. cDNA~Strawberry pyruvate
decarboxylase {Fragaria x ananassa}, partial (30%)
Length = 700
Score = 401 bits (1030), Expect(2) = e-115
Identities = 198/202 (98%), Positives = 200/202 (98%)
Frame = +1
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGI 60
MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGI
Sbjct: 49 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGI 228
Query: 61 TDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGL 120
TDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGG+
Sbjct: 229 TDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGV 408
Query: 121 SVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVV 180
SVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVV
Sbjct: 409 SVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVV 588
Query: 181 NNLEDAHEMIDTAISTALKESK 202
NNLEDAHEMIDTAISTALK +
Sbjct: 589 NNLEDAHEMIDTAISTALKRKQ 654
Score = 32.7 bits (73), Expect(2) = e-115
Identities = 14/15 (93%), Positives = 15/15 (99%)
Frame = +2
Query: 199 KESKPVYISISCNLA 213
KESKPVYISISCNL+
Sbjct: 644 KESKPVYISISCNLS 688
>BQ146924 similar to GP|10732644|gb pyruvate decarboxylase 1 {Vitis
vinifera}, partial (28%)
Length = 563
Score = 275 bits (703), Expect = 4e-74
Identities = 133/166 (80%), Positives = 148/166 (89%)
Frame = +3
Query: 122 VINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVN 181
V+NAIAGAYSENLP+ICI GGPNSND+G+ RILHHTIG+ DFSQEL+CFQTVTC+QAVVN
Sbjct: 66 VLNAIAGAYSENLPLICIXGGPNSNDYGSXRILHHTIGISDFSQELKCFQTVTCFQAVVN 245
Query: 182 NLEDAHEMIDTAISTALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMGLEA 241
+LEDAHE+IDTAISTALKESKPVYISISCNL +IPHPTFSREP+PFS +PKL+N MGLEA
Sbjct: 246 HLEDAHELIDTAISTALKESKPVYISISCNLPAIPHPTFSREPIPFSRAPKLTNQMGLEA 425
Query: 242 AVEAAAEVLNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVM 287
VEAAAE LNKAVKPVL GPKL AKA AF+EL D S YP +VM
Sbjct: 426 XVEAAAEFLNKAVKPVLXGGPKLXXAKASNAFVELXDASGYPLAVM 563
>TC87534 similar to SP|P17597|ILVB_ARATH Acetolactate synthase chloroplast
precursor (EC 4.1.3.18) (Acetohydroxy-acid synthase)
(ALS)., partial (87%)
Length = 2258
Score = 85.1 bits (209), Expect = 7e-17
Identities = 117/539 (21%), Positives = 213/539 (38%), Gaps = 22/539 (4%)
Frame = +2
Query: 19 ITTPTSNGTVSTIQKSPSTQSL---ASSESTLGSH-LARRLVEVGITDIFTVPGDFNLTL 74
I+ SN T T Q + A +E G+ L L G+T++F PG ++ +
Sbjct: 209 ISASVSNPTQKTTPIPSPEQFISRFAPNEPRKGADILVESLERQGVTNVFAYPGGASMEI 388
Query: 75 LDHLIAEPKLKNIGCCNELNAGYAADGYARSRGV-GACVVTFTVGGLSVINAIAGAYSEN 133
L ++N+ +E +AA+GYARS G+ G C+ T G ++++ +A A ++
Sbjct: 389 HQALTRSTAIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADAMLDS 568
Query: 134 LPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNNLEDAHEMIDTA 193
+P+I I G GT+ I + +++T + +V +++D ++ A
Sbjct: 569 VPLIAITGQVPRRMIGTDAFQETPI--------VEVTRSITKHNYLVLDVDDIPRIVKEA 724
Query: 194 ISTALK-ESKPVYISISCNL-ASIPHPTFSREPVPFSLSPKLSNPMGLEAAVEAAAEVLN 251
A PV I I ++ + P + +P+ + EA +E +L
Sbjct: 725 FLLASSGRPGPVLIDIPKDIQQQVSLPNWD-QPIRLTGYMNRLPKAPDEAHLEQIVRLLL 901
Query: 252 KAVKPVL-VAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVS 310
++ KPVL V G L ++ F+EL + P + G P ++ + G
Sbjct: 902 ESKKPVLYVGGGSLNCSEELRRFVEL---TGVPVASTLMGLGSYPSLDENSL-QMLGMHG 1069
Query: 311 TAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIGNGP---TFGCV 367
T + V+ +D L G F+D + + K + + D IG C
Sbjct: 1070TVYANYAVDKSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDAAEIGKNKQPHVSVCG 1249
Query: 368 LMKDFLSALAKRLKRNNTAYE----------NYFRIFVPEGLPVKSEPREPLRVNVLFQH 417
+K L + + L+ N + N ++ P E P Q
Sbjct: 1250DLKLALKGINQILESNGIERKLDFGGWREELNDQKVRFPMSYKTFDEAIPP---QYAIQV 1420
Query: 418 IQNMLSSETAVIAETGD-SWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEK 476
+ + + E + G + Q + + G++G+ + A +G A A P+
Sbjct: 1421LDELTNGEAIISTGVGQHQMWAAQFYRYKRPRQRLTSAGLGAMGFGLPAAMGAAVANPDA 1600
Query: 477 RVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG 535
V+ GDGSF + Q+++T+ I L+NN + V+ D Y + Y G
Sbjct: 1601IVVDIDGDGSFMMNVQELATIKVENLPVKILLLNNQHLGMVVQWEDRFYKANRAHTYLG 1777
>TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyase-like
protein - Arabidopsis thaliana, partial (75%)
Length = 1592
Score = 62.0 bits (149), Expect = 6e-10
Identities = 110/499 (22%), Positives = 193/499 (38%), Gaps = 52/499 (10%)
Frame = +1
Query: 52 ARRLVEVGITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAAD--GYARSRGVG 109
A+ + G+ +F V G +T L ++ I NE +AGYAA GY SR
Sbjct: 208 AKSFSQFGVLHMFGVVG-IPVTSLATRAVSLGIRFIAFHNEQSAGYAASAYGYLTSRPG- 381
Query: 110 ACVVTFTVGGLSVINAIAG---AYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQE 166
+ TV G ++ +AG + P + I G N ND +G DF QE
Sbjct: 382 ---IFLTVSGPGCVHGLAGLSNGTTNTWPTVMISGSCNQND----------VGRGDF-QE 519
Query: 167 LRCFQTVTCYQAVVNNLEDAHEMIDTAISTALKES---KP--VYISISCNLASIPHPTFS 221
L Q V + + E I ++ L S +P VY+ + ++ H S
Sbjct: 520 LDQIQAVKPFTKFAIKAKHISE-IPNCVAQVLANSVVNRPGGVYLDLPTDVL---HQKVS 687
Query: 222 REPVPFSLSP--------KLSNPMGLEAA-VEAAAEVLNKAVKPVLVAGPKLRVAKACEA 272
L+ K S+ + +E++ ++ A +L KA +P++V G AKA +
Sbjct: 688 ESEAESLLTEAKYLAEKIKKSHVISVESSKIQEAVSLLRKAERPLIVFGKGAAYAKAEDE 867
Query: 273 FIELADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVSTAFCAEIVESADAYLFAGPIFN 332
+L +K+ P+ P KGL+P+DH ++A + + D + G N
Sbjct: 868 LKKLVEKTGIPFLPTPMGKGLLPDDHP--------LAASAARSLAIGKCDVAIVIGARLN 1023
Query: 333 DYSSVGYS-----------LLLKKEKAIIVQPDRVVIGNGPTFGCVLMKD---------- 371
G + + KE+ + +P ++G+G VL K+
Sbjct: 1024WLLHFGEEPKWSKDVKFVLVDVSKEEIELRKPFLGLVGDGKEVLEVLNKEIKDDPFCLGS 1203
Query: 372 ---FLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHIQNMLSSETAV 428
++ A++ ++K N E V V P+R ++ I S V
Sbjct: 1204THPWVEAISSKVKDNGVKMEAQLAKDV-----VPFNFLTPMR--IIRDAISEFGGSPAPV 1362
Query: 429 IAETGDSWFNC---------QKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVI 479
+ G + + + +L G +G++G +G + A A P++ V+
Sbjct: 1363VVSEGANTMDVGRSVLVQKEPRTRLDAG-------TWGTMGVGLGYCIAAAVAYPDRLVV 1521
Query: 480 ACIGDGSFQVTAQDVSTML 498
A GD + +A +V +
Sbjct: 1522AVEGDSGYGFSAMEVEVYI 1578
>BQ140961 similar to SP|P09342|ILV1 Acetolactate synthase I chloroplast
precursor (EC 4.1.3.18) (Acetohydroxy-acid synthase I)
(ALS I)., partial (17%)
Length = 466
Score = 60.5 bits (145), Expect = 2e-09
Identities = 36/121 (29%), Positives = 63/121 (51%), Gaps = 2/121 (1%)
Frame = +1
Query: 23 TSNGTVSTIQKSPSTQSLASSESTLGSH-LARRLVEVGITDIFTVPGDFNLTLLDHLIAE 81
T+ + +T P T +S++ GS L L G+T++F PG ++ + L
Sbjct: 91 TAPPSTTTTVDEPFTSRFSSTQPRKGSDILVEALEREGVTNVFAYPGGASMEIHQALTRS 270
Query: 82 PKLKNIGCCNELNAGYAADGYARSRGV-GACVVTFTVGGLSVINAIAGAYSENLPVICIV 140
++NI +E +AA+GYARS G+ G C+ T G ++ + +A A +++P+I I
Sbjct: 271 KTIRNILPRHEQGGVFAAEGYARSSGLPGVCIATSGPGATNLXSGLADALMDSVPLIAIT 450
Query: 141 G 141
G
Sbjct: 451 G 453
>BQ750869 weakly similar to GP|21205303|db alpha-acetolactate synthase
{Staphylococcus aureus subsp. aureus MW2}, partial (18%)
Length = 732
Score = 50.4 bits (119), Expect = 2e-06
Identities = 28/88 (31%), Positives = 45/88 (50%), Gaps = 1/88 (1%)
Frame = +1
Query: 55 LVEVGITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRG-VGACVV 113
L+ G+ IF VPG ++ + LI P++K + C +E NA + A R G G C+
Sbjct: 88 LIAAGVKHIFGVPGAKIDSVFNALIDRPEIKLVVCRHEQNAAFIAGAIGRITGRPGVCIA 267
Query: 114 TFTVGGLSVINAIAGAYSENLPVICIVG 141
T G +++ + A E PV+ +VG
Sbjct: 268 TSGPGTSNLVTGLVTANDEGAPVVALVG 351
>TC84946 similar to GP|16245|emb|CAA35887.1|| precursor acetolactate
synthase (670 AA) {Arabidopsis thaliana}, partial (26%)
Length = 558
Score = 40.4 bits (93), Expect = 0.002
Identities = 24/79 (30%), Positives = 40/79 (50%)
Frame = +3
Query: 457 GSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTI 516
G++G+ + A +G A A P+ V+ GDGSF + Q+++T+ I L+NN +
Sbjct: 246 GAMGFGLPAAIGAAVANPDAIVVDIDGDGSFMMNVQELATIRVENLPVKILLLNNQHLGM 425
Query: 517 EVEIHDGPYNVIKNWNYTG 535
V+ D Y + Y G
Sbjct: 426 VVQWEDRFYKANRAHTYLG 482
>TC89160 similar to GP|9759326|dbj|BAB09835.1 contains similarity to
lysophosphatidic acid acyltransferase~gene_id:MUP24.4
{Arabidopsis thaliana}, partial (77%)
Length = 1433
Score = 31.6 bits (70), Expect = 0.92
Identities = 25/90 (27%), Positives = 38/90 (41%)
Frame = +1
Query: 394 FVPEGLPVKSEPREPLRVNVLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQ 453
++P G ++ EPR LR++ L I LS I + DS+ C K P+ +
Sbjct: 205 YLPSGAAIQQEPRGKLRLHDLLD-ISPTLSEAAGAIVD--DSFTRCFKSNPPEPWNWNI- 372
Query: 454 MQYGSIGWSVGATLGYAQAVPEKRVIACIG 483
Y W G L Y P + ++ IG
Sbjct: 373 --YLFPLWCFGVLLRYLIVFPTRVLVLTIG 456
>BQ752272 GP|1742479|dbj ORF_ID:o302#2~similar to {SwissProt Accession Number
P32051} [Escherichia coli], partial (53%)
Length = 1021
Score = 31.2 bits (69), Expect = 1.2
Identities = 17/51 (33%), Positives = 28/51 (54%)
Frame = -3
Query: 207 SISCNLASIPHPTFSREPVPFSLSPKLSNPMGLEAAVEAAAEVLNKAVKPV 257
++S ++AS+P PVPF P +S+P + +A LNK V+P+
Sbjct: 974 ALSPSIASVP-------PVPFRFPPPVSSP*LIPPLFTVSAPSLNKTVEPI 843
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 29.6 bits (65), Expect = 3.5
Identities = 14/38 (36%), Positives = 24/38 (62%)
Frame = +3
Query: 231 PKLSNPMGLEAAVEAAAEVLNKAVKPVLVAGPKLRVAK 268
PK +NP+ L A + AE ++++ KP++ PK + AK
Sbjct: 2511 PKFANPVLLSADIRKKAEAVDRSCKPIMTK-PKPKPAK 2621
>AW736467 similar to PIR|T05689|T0 hypothetical protein F20M13.170 -
Arabidopsis thaliana, partial (15%)
Length = 663
Score = 28.9 bits (63), Expect = 6.0
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = -2
Query: 391 FRIFVPEGLPVKSEPREPLRVNVLFQHIQNMLSS 424
F++F+ LP + R+ + VL Q +QN LSS
Sbjct: 482 FKLFITVALPTAIDNRDAAPMTVLVQKLQNALSS 381
>TC81198 similar to GP|21553392|gb|AAM62485.1 unknown {Arabidopsis
thaliana}, partial (38%)
Length = 913
Score = 28.5 bits (62), Expect = 7.8
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 2/30 (6%)
Frame = +3
Query: 194 ISTALKESKPVY--ISISCNLASIPHPTFS 221
IST+ K S P+Y + CNL P PTFS
Sbjct: 807 ISTSSKTSCPMYHLAYLECNLLLFPFPTFS 896
>BG448727 similar to PIR|T08924|T08 hypothetical protein T15N24.30 -
Arabidopsis thaliana, partial (31%)
Length = 684
Score = 28.5 bits (62), Expect = 7.8
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 2/30 (6%)
Frame = +1
Query: 194 ISTALKESKPVY--ISISCNLASIPHPTFS 221
IST+ K S P+Y + CNL P PTFS
Sbjct: 292 ISTSSKTSCPMYHLAYLECNLLLFPFPTFS 381
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,506,835
Number of Sequences: 36976
Number of extensions: 272832
Number of successful extensions: 1320
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 1309
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1319
length of query: 607
length of database: 9,014,727
effective HSP length: 102
effective length of query: 505
effective length of database: 5,243,175
effective search space: 2647803375
effective search space used: 2647803375
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140721.11


