

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.1 - phase: 0
(313 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
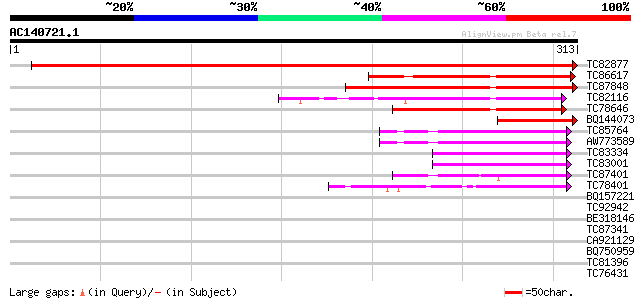
Score E
Sequences producing significant alignments: (bits) Value
TC82877 similar to GP|3335374|gb|AAC27175.1| expressed protein {... 612 e-176
TC86617 similar to PIR|C85062|C85062 probable thioredoxin [impor... 107 8e-24
TC87848 similar to GP|14326510|gb|AAK60300.1 AT3g54900/F28P10_12... 103 7e-23
TC82116 similar to GP|22165075|gb|AAM93692.1 putative PKCq-inter... 96 1e-20
TC78646 similar to GP|11994338|dbj|BAB02297. contains similarity... 79 2e-15
BQ144073 similar to GP|14326510|gb| AT3g54900/F28P10_120 {Arabid... 57 1e-08
TC85764 similar to GP|15637350|gb|AAL04507.1 glutaredoxin {Tilia... 47 9e-06
AW773589 similar to GP|10178147|db glutaredoxin-like protein {Ar... 47 9e-06
TC83334 weakly similar to GP|21618084|gb|AAM67134.1 glutaredoxin... 43 2e-04
TC83001 similar to GP|21618084|gb|AAM67134.1 glutaredoxin-like p... 42 3e-04
TC87401 similar to PIR|B84587|B84587 probable glutaredoxin [impo... 41 5e-04
TC78401 similar to GP|6735386|emb|CAB69043.1 glutaredoxin {Arabi... 41 7e-04
BQ157221 similar to GP|21617983|gb| glutaredoxin-like protein {A... 39 0.003
TC92942 similar to GP|15451042|gb|AAK96792.1 putative protein {A... 37 0.008
BE318146 similar to GP|17064874|gb| Unknown protein {Arabidopsis... 35 0.037
TC87341 similar to PIR|T12219|T12219 glutaredoxin I - common ice... 35 0.037
CA921129 similar to GP|14532460|gb AT5g13810/MAC12_24 {Arabidops... 34 0.064
BQ750959 similar to GP|18376329|emb related to cell surface ferr... 32 0.41
TC81396 similar to PIR|T06535|T06535 heat shock transcription fa... 32 0.41
TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 31 0.70
>TC82877 similar to GP|3335374|gb|AAC27175.1| expressed protein {Arabidopsis
thaliana}, partial (70%)
Length = 1158
Score = 612 bits (1578), Expect = e-176
Identities = 300/301 (99%), Positives = 301/301 (99%)
Frame = +2
Query: 13 DHSSMATIINLSSTQLLSYHHFPSFSPQNTPTLSLNFHPNSSTNLRPISLKPYHPRKPQS 72
+HSSMATIINLSSTQLLSYHHFPSFSPQNTPTLSLNFHPNSSTNLRPISLKPYHPRKPQS
Sbjct: 5 EHSSMATIINLSSTQLLSYHHFPSFSPQNTPTLSLNFHPNSSTNLRPISLKPYHPRKPQS 184
Query: 73 WLVAMAVKNLADTSPVTVSPENDGLTGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVL 132
WLVAMAVKNLADTSPVTVSPENDGLTGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVL
Sbjct: 185 WLVAMAVKNLADTSPVTVSPENDGLTGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVL 364
Query: 133 AHRKSVPELCGSFKVGVVDEPDRESLTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQP 192
AHRKSVPELCGSFKVGVVDEPDRESLTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQP
Sbjct: 365 AHRKSVPELCGSFKVGVVDEPDRESLTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQP 544
Query: 193 KKKADLRLTPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEG 252
KKKADLRLTPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEG
Sbjct: 545 KKKADLRLTPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEG 724
Query: 253 VDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
VDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK
Sbjct: 725 VDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 904
Query: 313 K 313
K
Sbjct: 905 K 907
>TC86617 similar to PIR|C85062|C85062 probable thioredoxin [imported] -
Arabidopsis thaliana, partial (49%)
Length = 903
Score = 107 bits (266), Expect = 8e-24
Identities = 55/114 (48%), Positives = 76/114 (66%)
Frame = +2
Query: 199 RLTPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESV 258
++ PG L L+ELVD + V+ F+KGS P CGFS+KV+ IL++E V + S
Sbjct: 473 KVQPGLSSHLKKRLQELVDS----HPVLLFMKGSPEEPKCGFSRKVVDILKEEKVKFGSF 640
Query: 259 DVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
D+L + +RE LKK+SNWPTFPQ++ GELVGGCDI +M+E GE+ +FK
Sbjct: 641 DILSDSE---VREGLKKFSNWPTFPQLYCKGELVGGCDIAIAMHESGELKDVFK 793
>TC87848 similar to GP|14326510|gb|AAK60300.1 AT3g54900/F28P10_120
{Arabidopsis thaliana}, partial (63%)
Length = 772
Score = 103 bits (258), Expect = 7e-23
Identities = 54/129 (41%), Positives = 79/129 (60%), Gaps = 1/129 (0%)
Frame = +3
Query: 186 TWVRPQPKKKADLRL-TPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKV 244
T + QPK + R T R LT L+ +DK+V NK+V F+KG++ P CGFS V
Sbjct: 198 TILHSQPKFQLKQRFSTTIRCSALTPELKTTLDKVVTSNKIVLFMKGTKDFPQCGFSNTV 377
Query: 245 IGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEK 304
+ IL +E++++LD D LR+ LK+YS+WPTFPQ++++GE GGCDI +
Sbjct: 378 VQILRSLNASFETINILDNDM---LRQGLKEYSSWPTFPQLYIDGEFFGGCDITVEAYKN 548
Query: 305 GEVAGLFKK 313
GE+ L +K
Sbjct: 549 GELQELVEK 575
>TC82116 similar to GP|22165075|gb|AAM93692.1 putative PKCq-interacting
protein {Oryza sativa (japonica cultivar-group)},
partial (33%)
Length = 692
Score = 96.3 bits (238), Expect = 1e-20
Identities = 58/165 (35%), Positives = 96/165 (58%), Gaps = 6/165 (3%)
Frame = +3
Query: 149 VVDEPDRESLT--QAWKSWMEEHIKITGKVPPGNESGNATWVRPQPKKKADLRLTPGRHV 206
+ DE R+ L W S+ + +IK G++ G++ + + +K +L+ T H
Sbjct: 36 LTDEEVRQGLKVYSNWSSYPQLYIK--GELIGGSD------IVLEMQKSGELQKT--LHE 185
Query: 207 QLTVPLEELVD---KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGV-DYESVDVLD 262
+ +P E + D KL+ + V+ F+KG+ AP CGFS +V+ L +EGV D+ D+L
Sbjct: 186 KGVLPKETIEDRLKKLIASSPVMLFMKGTPDAPRCGFSSRVVNALREEGVVDFGHFDILS 365
Query: 263 EDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+D +R+ +K +SNWPTFPQ++ GEL+GGCDI+ + GE+
Sbjct: 366 DDE---VRQGIKVFSNWPTFPQLYYKGELIGGCDIIMELRNNGEL 491
>TC78646 similar to GP|11994338|dbj|BAB02297. contains similarity to
thioredoxin~gene_id:MSJ11.6 {Arabidopsis thaliana},
partial (68%)
Length = 807
Score = 79.3 bits (194), Expect = 2e-15
Identities = 34/96 (35%), Positives = 63/96 (65%)
Frame = +1
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
L ++++ VK+N V+ ++KG P CGFS + +L++ V + ++L + +++
Sbjct: 199 LSSIIEQDVKDNPVMIYMKGVPDFPQCGFSSLAVKVLKQYDVPLSARNILQDPE---VKD 369
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+K +S+WPTFPQ+F+ GE +GG DI+ SM++ GE+
Sbjct: 370 AVKAFSHWPTFPQVFIKGEFIGGSDIVLSMHQSGEL 477
>BQ144073 similar to GP|14326510|gb| AT3g54900/F28P10_120 {Arabidopsis
thaliana}, partial (30%)
Length = 475
Score = 56.6 bits (135), Expect = 1e-08
Identities = 23/44 (52%), Positives = 32/44 (72%)
Frame = +3
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
RE LK+YS+WPTFPQ++++GE GGCDI + GE+ L +K
Sbjct: 66 REGLKEYSSWPTFPQLYIDGEFFGGCDITVEAYKNGELQELVEK 197
>TC85764 similar to GP|15637350|gb|AAL04507.1 glutaredoxin {Tilia
platyphyllos}, partial (87%)
Length = 734
Score = 47.0 bits (110), Expect = 9e-06
Identities = 27/106 (25%), Positives = 53/106 (49%)
Frame = +2
Query: 205 HVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDED 264
H ++ +P + ++V N VV F K C F +V + GV +++V++ E
Sbjct: 131 HSKMALPKAK---EIVSSNPVVVFSKS-----YCPFCVQVKKLFTDLGVTFKAVELDSES 286
Query: 265 YNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
++ L +++ T P +F+ G +GGCD T++ +G++ L
Sbjct: 287 DGSEIQGALAQWTGQRTVPNVFIGGNHIGGCDSTTNLQNQGKLVPL 424
>AW773589 similar to GP|10178147|db glutaredoxin-like protein {Arabidopsis
thaliana}, complete
Length = 543
Score = 47.0 bits (110), Expect = 9e-06
Identities = 27/106 (25%), Positives = 53/106 (49%)
Frame = -1
Query: 205 HVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDED 264
H ++ +P + ++V N VV F K C F +V + GV +++V++ E
Sbjct: 420 HSKMALPKAK---EIVSSNPVVVFSKS-----YCPFCVQVKKLFTDLGVTFKAVELDSES 265
Query: 265 YNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
++ L +++ T P +F+ G +GGCD T++ +G++ L
Sbjct: 264 DGSEIQGALAQWTGQRTVPNVFIGGNHIGGCDSTTNLQNQGKLVPL 127
>TC83334 weakly similar to GP|21618084|gb|AAM67134.1 glutaredoxin-like
protein {Arabidopsis thaliana}, partial (91%)
Length = 603
Score = 42.7 bits (99), Expect = 2e-04
Identities = 19/77 (24%), Positives = 41/77 (52%)
Frame = +2
Query: 234 SAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVG 293
S CG+ ++V +L++ G Y+ +++ E + L +++ T P +F+ G+ +G
Sbjct: 182 SKTYCGYCKRVKDLLKQLGATYKVLEMDIESDGDEIHAALTEWTGQRTVPNVFIGGKHIG 361
Query: 294 GCDILTSMNEKGEVAGL 310
GCD + + G++ L
Sbjct: 362 GCDSILEKHRAGQLIPL 412
>TC83001 similar to GP|21618084|gb|AAM67134.1 glutaredoxin-like protein
{Arabidopsis thaliana}, partial (86%)
Length = 641
Score = 42.0 bits (97), Expect = 3e-04
Identities = 18/77 (23%), Positives = 41/77 (52%)
Frame = +2
Query: 234 SAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVG 293
S CG+ +V +L++ G ++ +++ E ++ L +++ T P +F+ G+ +G
Sbjct: 122 SKTYCGYCNRVKDLLKQLGAAHKVIELDTESDGGEIQAALAEWTGQRTVPNVFIGGKHIG 301
Query: 294 GCDILTSMNEKGEVAGL 310
GCD + + G++ L
Sbjct: 302 GCDSVLEKHRTGQLVPL 352
>TC87401 similar to PIR|B84587|B84587 probable glutaredoxin [imported] -
Arabidopsis thaliana, partial (63%)
Length = 914
Score = 41.2 bits (95), Expect = 5e-04
Identities = 28/101 (27%), Positives = 49/101 (47%), Gaps = 2/101 (1%)
Frame = +3
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG--L 269
LEE + V +N VV + K C + +V + +K G ++ LDE G L
Sbjct: 279 LEETIKNTVSQNPVVVYSKS-----WCSYCSEVKSLFKKLGTQPLVIE-LDELGPQGPQL 440
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
++ L++ + T P +F+ G+ +GGC + KG++ L
Sbjct: 441 QKLLERITGQYTVPNVFIGGQHIGGCTDTLKLYRKGDLETL 563
>TC78401 similar to GP|6735386|emb|CAB69043.1 glutaredoxin {Arabidopsis
thaliana}, partial (68%)
Length = 706
Score = 40.8 bits (94), Expect = 7e-04
Identities = 41/146 (28%), Positives = 67/146 (45%), Gaps = 12/146 (8%)
Frame = +3
Query: 177 PPGNESGNATWVRPQPKKKADLRLTPGRHVQ---LTVPLE---------ELVDKLVKENK 224
PP ++S + R QP K A +++T + ++V L+ E VDK + NK
Sbjct: 9 PPYSQSSDR---RDQPFKSARMKVTMMMVMMVLAISVYLDSASAASSVGEFVDKTINNNK 179
Query: 225 VVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQ 284
+ F K P C ++ V L + V Y V++ + D +++ L T PQ
Sbjct: 180 IAIFSK--TYCPYCRRAKAVFKELNQ--VPYV-VELDERDDGSKIQDVLVNIVGKRTVPQ 344
Query: 285 IFLNGELVGGCDILTSMNEKGEVAGL 310
+F+NG+ +GG D E G +A L
Sbjct: 345 VFINGKHLGGSDETVEAYESGLLAKL 422
>BQ157221 similar to GP|21617983|gb| glutaredoxin-like protein {Arabidopsis
thaliana}, partial (94%)
Length = 672
Score = 38.5 bits (88), Expect = 0.003
Identities = 29/107 (27%), Positives = 51/107 (47%), Gaps = 1/107 (0%)
Frame = +3
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNY 267
LT+ + E V K+V E VV F K S C S + + GV+ ++ +
Sbjct: 54 LTLYIMERVTKMVSERPVVIFSKSS-----CCMSHTIKTLFCDFGVNPAVYELDEIPRGR 218
Query: 268 GLRETL-KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ + L + + P+ P +F+ GELVGG + + S++ + + KK
Sbjct: 219 EIEQALISRLGSSPSVPTVFIGGELVGGANQVMSLHLNRSLIPMLKK 359
>TC92942 similar to GP|15451042|gb|AAK96792.1 putative protein {Arabidopsis
thaliana}, partial (38%)
Length = 650
Score = 37.4 bits (85), Expect = 0.008
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 8/110 (7%)
Frame = +1
Query: 211 PLEELVDKLV-----KENKVVAF---IKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLD 262
P L DK + EN++V + ++G R + ++I + VD + +D
Sbjct: 34 PCSSLFDKSLCLLPGTENRIVVYCTSLRGIRKTYEDCCAVRMILRGYRVAVDERDIS-MD 210
Query: 263 EDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
Y L+ L S T PQ+F+ G+ VG D L +NE GE+A + K
Sbjct: 211 SSYRKELQNALGGKSV-VTLPQVFIRGKHVGNADDLKQLNESGELARMLK 357
>BE318146 similar to GP|17064874|gb| Unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 382
Score = 35.0 bits (79), Expect = 0.037
Identities = 28/82 (34%), Positives = 45/82 (54%), Gaps = 4/82 (4%)
Frame = +3
Query: 6 LLLTHSTDHSSMA-TIINLSSTQLLSYHHFPSFSP---QNTPTLSLNFHPNSSTNLRPIS 61
LLL+ +T+ S + + IN+ S+ + +HH+P P T TL+L PN+S P S
Sbjct: 27 LLLSLTTNISFLCFSFINIHSSSSIPFHHYPLLLPFFLSLTFTLTL---PNAS----PRS 185
Query: 62 LKPYHPRKPQSWLVAMAVKNLA 83
HPR+ +SW + ++ LA
Sbjct: 186 NHNSHPRERESWSLCVSQMWLA 251
>TC87341 similar to PIR|T12219|T12219 glutaredoxin I - common ice plant,
partial (73%)
Length = 787
Score = 35.0 bits (79), Expect = 0.037
Identities = 26/97 (26%), Positives = 47/97 (47%)
Frame = +2
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
V + N++ F K P C +++V L ++ E +D+ D+ Y ++ L
Sbjct: 197 VQNAIYSNRITIFSKSY--CPYCLRAKRVFVELNEQPFVIE-LDLRDDGYQ--IQGVLLD 361
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
T PQ+F+ G+ +GG D L++ + GE+ L K
Sbjct: 362 LIGRRTVPQVFVYGKHIGGSDDLSAAVQSGELQKLLK 472
>CA921129 similar to GP|14532460|gb AT5g13810/MAC12_24 {Arabidopsis
thaliana}, partial (40%)
Length = 729
Score = 34.3 bits (77), Expect = 0.064
Identities = 12/27 (44%), Positives = 20/27 (73%)
Frame = -2
Query: 281 TFPQIFLNGELVGGCDILTSMNEKGEV 307
T PQ+F+ G+ +GG D++ S+ E GE+
Sbjct: 611 TLPQVFVRGKYIGGADVIKSLFETGEL 531
>BQ750959 similar to GP|18376329|emb related to cell surface ferroxidase
{Neurospora crassa}, partial (13%)
Length = 663
Score = 31.6 bits (70), Expect = 0.41
Identities = 17/43 (39%), Positives = 22/43 (50%)
Frame = +1
Query: 61 SLKPYHPRKPQSWLVAMAVKNLADTSPVTVSPENDGLTGEELP 103
S+ PYH P+ A +KN+A T P+ V P G TG P
Sbjct: 148 SMAPYHEMTPRPSKAATTLKNMA-T*PLAVKPSGSGPTGPFSP 273
>TC81396 similar to PIR|T06535|T06535 heat shock transcription factor A -
garden pea (fragment), partial (91%)
Length = 1138
Score = 31.6 bits (70), Expect = 0.41
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 13/82 (15%)
Frame = +3
Query: 152 EPDRESLTQAWKSWMEEHIK---ITGKVPPGNES--GNATW--------VRPQPKKKADL 198
E + E+L A+ + +K VP GNES G+A W V P+ + +
Sbjct: 900 ESEMETLLSAYDNESSSEVKDYTALSSVPTGNESNLGDAVWEDLLNQELVGGNPEDEVVI 1079
Query: 199 RLTPGRHVQLTVPLEELVDKLV 220
G Q+ VP+EELV+K++
Sbjct: 1080----GDFSQIDVPVEELVEKMI 1133
>TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (30%)
Length = 890
Score = 30.8 bits (68), Expect = 0.70
Identities = 17/57 (29%), Positives = 26/57 (44%), Gaps = 10/57 (17%)
Frame = +2
Query: 14 HSSMATIINLSSTQLLSYHHFPSFSPQN----------TPTLSLNFHPNSSTNLRPI 60
H T I L+ TQ ++HH + S + +PT +L+ H S TN P+
Sbjct: 143 HHHQPTTITLTPTQFTTHHHHQNTSIPHHHHQCITSALSPTTTLHHHQRSHTNTHPL 313
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,702,004
Number of Sequences: 36976
Number of extensions: 166502
Number of successful extensions: 920
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 911
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 914
length of query: 313
length of database: 9,014,727
effective HSP length: 96
effective length of query: 217
effective length of database: 5,465,031
effective search space: 1185911727
effective search space used: 1185911727
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140721.1


