

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140671.7 - phase: 0
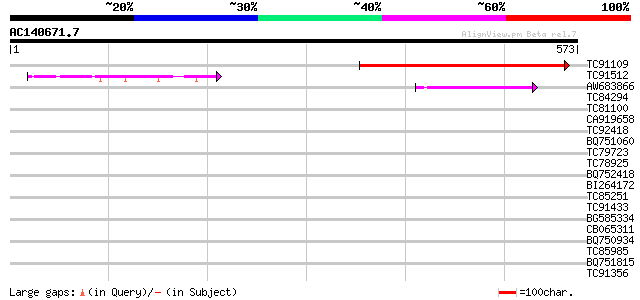
(573 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91109 similar to GP|15028145|gb|AAK76696.1 unknown protein {Ar... 238 4e-63
TC91512 similar to PIR|D85359|D85359 hypothetical protein AT4g30... 86 4e-17
AW683866 similar to GP|15217302|gb| Hypothetical protein {Oryza ... 74 2e-13
TC84294 homologue to GP|168765|gb|AAA33563.1|| plasma membrane H... 40 0.002
TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon prot... 39 0.004
CA919658 weakly similar to PIR|D85359|D853 hypothetical protein ... 37 0.021
TC92418 similar to GP|3551960|gb|AAC34858.1| senescence-associat... 36 0.035
BQ751060 similar to GP|14330324|emb aldo-keto reductase {Gallus ... 35 0.060
TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-al... 35 0.060
TC78925 similar to GP|17529158|gb|AAL38805.1 unknown protein {Ar... 35 0.060
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 35 0.078
BI264172 homologue to PIR|T45739|T457 transcription factor-like ... 35 0.078
TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean... 35 0.078
TC91433 35 0.078
BG585334 similar to GP|20161490|db beta-1 3 glucanase-like prote... 34 0.13
CB065311 similar to PIR|C83375|C83 probable glycosyl hydrolase P... 34 0.17
BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 ... 34 0.17
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 34 0.17
BQ751815 similar to PIR|S58652|S586 hypothetical protein YFR036w... 33 0.23
TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophil... 33 0.23
>TC91109 similar to GP|15028145|gb|AAK76696.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1166
Score = 238 bits (608), Expect = 4e-63
Identities = 130/213 (61%), Positives = 161/213 (75%), Gaps = 1/213 (0%)
Frame = +1
Query: 354 SPIRSAVRPASPSKLGTPSPRSPSRGVS-PCRGRNGVASSLSSRFVNEPSVLSFAVDVPR 412
SPIR RPASPSKL + SPSRG S P + R+ VAS+++S + PS+LSF+ DV R
Sbjct: 4 SPIRGNARPASPSKLWASAASSPSRGFSSPSKVRSAVASTINSNSGSSPSILSFSADVRR 183
Query: 413 GKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLRESV 472
GKIGE+R+ DAH+LRLL+NR +QWRFVNARADA+ VQ LN+E L+ AWV S+LR SV
Sbjct: 184 GKIGEDRIFDAHTLRLLYNRYVQWRFVNARADAAFMVQKLNAETHLWNAWVTISELRHSV 363
Query: 473 VAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPVFGG 532
+ KRI L LL+Q LKL SIL Q+ YLE+WA+LDR +S S+ GATEAL+ASTLRLP+
Sbjct: 364 ILKRIKLVLLRQKLKLTSILKGQISYLEEWALLDRDHSSSVLGATEALRASTLRLPLVEK 543
Query: 533 AKIDLLNLKEAICSAMDVMQAMASSICLLLPKV 565
A D+ NLK+A+ SA+DVMQAMASSI L KV
Sbjct: 544 ATADVPNLKDALGSAVDVMQAMASSIYSLSSKV 642
>TC91512 similar to PIR|D85359|D85359 hypothetical protein AT4g30710
[imported] - Arabidopsis thaliana, partial (23%)
Length = 1285
Score = 85.9 bits (211), Expect = 4e-17
Identities = 83/247 (33%), Positives = 118/247 (47%), Gaps = 51/247 (20%)
Frame = +2
Query: 19 TPTRPPLLPSESDNAI-VRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPL-VNRTVNST 76
TP RP L+P+E +NAI R+ REV+SRY S + + +S P+R SP +R + +
Sbjct: 341 TPRRP-LVPAEKNNAIPTRRSGTREVSSRY---KSPTPAAASGPQRCPSPSGTHRCPSPS 508
Query: 77 KQKSTPSVTPAAFV----------KRSQSTERQRQRQGTPRPNGETPVA----------- 115
+ PS P++F KR+ S ER+R T P+ TP
Sbjct: 509 GPRRCPS--PSSFTRTTAASKLLPKRALSAERKRPTTPTSPPSPSTPAQDDVHLSRRAAS 682
Query: 116 ---QKMLFTSA-RSLSVSFQGESFSIPVSKAKPPSATV-----------------TQSLR 154
Q+ L+ S RSLSVSFQ ++ SIPVSK + P + T + R
Sbjct: 683 SRMQESLWPSTMRSLSVSFQSDTISIPVSKKERPVTSASDRTLRPTSNVAHKQVETPNTR 862
Query: 155 RSTPERRKVPATVTTPTIGRGRVNGNSDQTENS------ISRSGDQHRWPAK-SQQNQAN 207
+ TPERR+ P + SDQ+ENS SR DQHRWP++ + +N
Sbjct: 863 KPTPERRRSPL----------KGKNASDQSENSKPVDGLQSRLIDQHRWPSRIGGKVSSN 1012
Query: 208 FMNRSLD 214
+NRS+D
Sbjct: 1013SLNRSVD 1033
>AW683866 similar to GP|15217302|gb| Hypothetical protein {Oryza sativa}
[Oryza sativa (japonica cultivar-group)], partial (4%)
Length = 552
Score = 73.6 bits (179), Expect = 2e-13
Identities = 41/123 (33%), Positives = 64/123 (51%)
Frame = +3
Query: 411 PRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLRE 470
P G N D H LRLL NRL+QWR+ NA A + + ++E +L W +K R
Sbjct: 69 PSNGFGNNE--DVHKLRLLDNRLIQWRYANAXAHIVNANISRHTESNLICVWDGLTKSRN 242
Query: 471 SVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPVF 530
+V+ K+I K +K IL Q+ LE W ++R + +++ E L ++ R+P+
Sbjct: 243 TVMKKKIQFAREKLXMKKAFILYYQLKLLEAWGSMERQHVSTITATKECLHSAVCRIPLL 422
Query: 531 GGA 533
GA
Sbjct: 423 EGA 431
>TC84294 homologue to GP|168765|gb|AAA33563.1|| plasma membrane H+ ATPase
{Neurospora crassa}, partial (29%)
Length = 815
Score = 40.4 bits (93), Expect = 0.002
Identities = 37/118 (31%), Positives = 54/118 (45%)
Frame = +2
Query: 7 TTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNS 66
+T ++ ++ P+P R P PS + + R P +R + S SS SS + PRR+
Sbjct: 242 STRSSSSSPSTPSPRRLP--PSSNPPTVRRLPASRVLPS---LSSRPSSRTTLSPRRSTR 406
Query: 67 PLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSAR 124
P R +S S PSV PA+ V +T R P P TP+A + S R
Sbjct: 407 PTRTRLPSSPPVVSAPSVLPASVV----TTVPGRFSVSCPAP---TPLATTLPAPSTR 559
>TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana, partial (20%)
Length = 1266
Score = 39.3 bits (90), Expect = 0.004
Identities = 47/167 (28%), Positives = 76/167 (45%), Gaps = 12/167 (7%)
Frame = +3
Query: 14 TKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTV 73
T R+ TP+ P LPS + ++ P +R ++ +S+ ++PP R+++P
Sbjct: 747 TSRSSTPSGRPTLPSTTKSSRPSTPTSR--------ATLTSTKSTAPPVRSSTP-----T 887
Query: 74 NSTKQKSTPSVTPAAFV-KRSQSTERQRQRQGTP-RPNG-ETPVAQKMLFTSARSLSVSF 130
T + STP+ P+ K SQ + R TP R G P + + AR +
Sbjct: 888 RPTSRASTPTSRPSLTAPKTSQRSATPNIRSSTPSRTFGVSAPPTRPSSTSKARPVVAKN 1067
Query: 131 QGESFSI-PVSKAKP------PSAT--VTQSLRRSTPERRKVPATVT 168
+S I P K++P P T +L++S PER PA+VT
Sbjct: 1068PVQSRGISPSVKSRPWEPSQMPGFTHDAPSNLKKSLPER---PASVT 1199
>CA919658 weakly similar to PIR|D85359|D853 hypothetical protein AT4g30710
[imported] - Arabidopsis thaliana, partial (13%)
Length = 809
Score = 37.0 bits (84), Expect = 0.021
Identities = 23/50 (46%), Positives = 33/50 (66%), Gaps = 5/50 (10%)
Frame = -1
Query: 512 SLSGATEALKASTLRLPVFGGAKI----DLLNLKEAI-CSAMDVMQAMAS 556
+LSGA E L+A+TLRLP+ GGAKI LL+++ + C D+ A+ S
Sbjct: 800 ALSGAVEDLEANTLRLPLTGGAKILSP*RLLSVQLLMSCKLWDLQSALCS 651
Score = 34.3 bits (77), Expect = 0.13
Identities = 18/30 (60%), Positives = 22/30 (73%)
Frame = -3
Query: 536 DLLNLKEAICSAMDVMQAMASSICLLLPKV 565
D+ LK AICSA+DVMQAM S+I L +V
Sbjct: 732 DIEPLKVAICSAVDVMQAMGSAIRPLFSRV 643
>TC92418 similar to GP|3551960|gb|AAC34858.1| senescence-associated protein
15 {Hemerocallis hybrid cultivar}, partial (35%)
Length = 1034
Score = 36.2 bits (82), Expect = 0.035
Identities = 19/41 (46%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Frame = +3
Query: 47 YMYSSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPS-VTP 86
+ +S SSS S+SS P +NSP+ N TV+ +TPS +TP
Sbjct: 222 FSFSHSSSLSYSSSPFSSNSPI*NSTVSHNSYSNTPSNLTP 344
>BQ751060 similar to GP|14330324|emb aldo-keto reductase {Gallus gallus},
partial (20%)
Length = 827
Score = 35.4 bits (80), Expect = 0.060
Identities = 37/161 (22%), Positives = 63/161 (38%), Gaps = 8/161 (4%)
Frame = +2
Query: 12 KNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYM--------YSSSSSSSFSSPPRR 63
++ + +PTP+R PS + A + ++ V+ R + +SSS S +PP
Sbjct: 224 RSPRPSPTPSRSATRPSTAPTATPTRTRSARVSRRPLPRASSVRTFSSSPSCGLPTPPAT 403
Query: 64 TNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSA 123
+ S V+R + + + F T R+ P P + + +
Sbjct: 404 SASRRVSR--RA*RAWDWTMLICTLFTGLLP*TPRETTTDSPPSPTEAVTFSATLTTSRP 577
Query: 124 RSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVP 164
S S S + S+P+ A P T T S+PE R P
Sbjct: 578 GSPSRSSSRRARSVPLVSATTPRNTSTS----SSPEARSSP 688
>TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-alpha
(EF-1-alpha). {Podospora anserina}, complete
Length = 1537
Score = 35.4 bits (80), Expect = 0.060
Identities = 40/165 (24%), Positives = 65/165 (39%), Gaps = 7/165 (4%)
Frame = +3
Query: 14 TKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSS-----SSSSSFSSPPRRTNSPL 68
+ R TP+ PP S + + R V S S+ SSS ++SSP SPL
Sbjct: 756 SSRLLTPSSPPSARPTSPSVSPSRMSTRSVVSALCPSAVSRLVSSSPAWSSPSLLPTSPL 935
Query: 69 VNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSV 128
+ T S + V +++ + T T +L ++ RSLS+
Sbjct: 936 KSSPSRCTTSSSLRASPVTMLVST*RTSPSRTFAVATSPVTPRTTPLPALLPSTLRSLSL 1115
Query: 129 SFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATV--TTPT 171
+ S + + T+ S RS+ VPA++ TTP+
Sbjct: 1116TTPVRSVLVTPRSSTATLPTLPASSPRSSRRSTAVPASLSRTTPS 1250
>TC78925 similar to GP|17529158|gb|AAL38805.1 unknown protein {Arabidopsis
thaliana}, partial (37%)
Length = 1274
Score = 35.4 bits (80), Expect = 0.060
Identities = 38/145 (26%), Positives = 56/145 (38%), Gaps = 2/145 (1%)
Frame = -2
Query: 4 AISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP-- 61
++ T T ++ P PL P + N + RKP + + SS+ SSS + P
Sbjct: 625 SLKLTPTHSPEQQPMQPQLQPLPPPSTANQVFRKPPYQNTQAISSLSSNHSSSTKTTPFH 446
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFT 121
+SP + N + ST S T R T +P TP++ T
Sbjct: 445 LSPDSPTPLSSYNLRRGCSTASYT-------ESDPGSHRVSYNTKKP---TPISVHSNQT 296
Query: 122 SARSLSVSFQGESFSIPVSKAKPPS 146
S V + S PVSK K P+
Sbjct: 295 QTHSDPV*TGFQHHSFPVSKPKSPA 221
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 35.0 bits (79), Expect = 0.078
Identities = 35/138 (25%), Positives = 56/138 (40%), Gaps = 13/138 (9%)
Frame = -2
Query: 4 AISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFS---SP 60
++ TT + +P+PT +LP A R+P++ +R S+S ++ S
Sbjct: 470 SLPTTRALASRSLSPSPTTRAVLPRRKSTAWSRRPRSTPRKTRLPASASRLATVSRTTPS 291
Query: 61 PRRTNSP----LVNRTVNSTKQK-STPSVTPAAFVKR-----SQSTERQRQRQGTPRPNG 110
P RT S L R+ T++ S PS P +R + T R R+R P
Sbjct: 290 PSRTRSTTRRVLAARSTTRTRRLFSRPSRRPTTGSRRTAPLPTPRTSRSRRRSCPTSPTP 111
Query: 111 ETPVAQKMLFTSARSLSV 128
P + L A S+
Sbjct: 110 SPPRCTRALVAPAARTSL 57
>BI264172 homologue to PIR|T45739|T457 transcription factor-like protein -
Arabidopsis thaliana, partial (4%)
Length = 568
Score = 35.0 bits (79), Expect = 0.078
Identities = 29/83 (34%), Positives = 43/83 (50%), Gaps = 4/83 (4%)
Frame = -1
Query: 17 APTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRT----NSPLVNRT 72
+P+P ++ E ++ VR EV + SSSSSSS +S T NSPL +
Sbjct: 463 SPSPAPENVVEFELSSSSVRSCSEPEVVLFFSCSSSSSSSQTSTSFPTEPC*NSPLEKSS 284
Query: 73 VNSTKQKSTPSVTPAAFVKRSQS 95
+ST +KS+P+ T AF + S
Sbjct: 283 KSST-EKSSPTTTLFAFKHKISS 218
>TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean, partial
(93%)
Length = 1554
Score = 35.0 bits (79), Expect = 0.078
Identities = 42/174 (24%), Positives = 63/174 (36%), Gaps = 4/174 (2%)
Frame = -2
Query: 3 TAISTTTTTKNTKRAPTPTRPPLL----PSESDNAIVRKPKAREVTSRYMYSSSSSSSFS 58
TA + T T P T+PP + P S + PK+ VTS + +SS
Sbjct: 842 TAPTKLPPTTPTAITPVTTQPPTVVASPPITSQPPVTVAPKSAPVTSPAPKIAPASSPKV 663
Query: 59 SPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKM 118
PP+ S V+ P ++P Q+ + TP P P K
Sbjct: 662 PPPQPPKSSPVSTPTLPPPLPPPPKISPTPV----QTPPAPAPVKATPVP---APAPAKQ 504
Query: 119 LFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTI 172
T A + S + +I P S+ + RR P+ R+ A PT+
Sbjct: 503 APTPAPATSPPIPAPTPAIEAPVPAPESSKPKR--RRHRPKHRRHQAPAPAPTV 348
>TC91433
Length = 738
Score = 35.0 bits (79), Expect = 0.078
Identities = 30/145 (20%), Positives = 58/145 (39%)
Frame = +2
Query: 14 TKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTV 73
T + T + P++ + + ++P A TS ++ ++++ S P T +P T+
Sbjct: 302 TPASNTNKQQPIVAATPASNTNKQPAATSPTSNKPITTVTAAAPSKLP--TTNPSAPSTL 475
Query: 74 NSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGE 133
KQK + TPA+ K+ + + + + P + +A S +
Sbjct: 476 PLDKQKPIVAATPASSTKQPPAATSPISNKPSTTISAAAPSKLPTIIPAAPSKLPTAASP 655
Query: 134 SFSIPVSKAKPPSATVTQSLRRSTP 158
+ S P + KPP T L P
Sbjct: 656 TSSTPSAAKKPPIGVATSPLANKQP 730
>BG585334 similar to GP|20161490|db beta-1 3 glucanase-like protein {Oryza
sativa (japonica cultivar-group)}, partial (39%)
Length = 835
Score = 34.3 bits (77), Expect = 0.13
Identities = 27/92 (29%), Positives = 41/92 (44%), Gaps = 15/92 (16%)
Frame = +1
Query: 10 TTKNTKRAPTPTRPPLLPSESDNAIV-RKPKAREV-----TSRYMYSSSSSSSFSSPPRR 63
TT T P PT P P D + + P + + T+ Y +S+S +S+PP
Sbjct: 160 TTPGTGGNPYPTINPTSPQPPDTSTGGQNPPSPDTDTTSPTNPYSNPPTSTSPYSNPPAS 339
Query: 64 TN---------SPLVNRTVNSTKQKSTPSVTP 86
TN +P N T +T ++P+VTP
Sbjct: 340 TNPYSNPPASTNPYSNPTSPTTTTPTSPTVTP 435
>CB065311 similar to PIR|C83375|C83 probable glycosyl hydrolase PA2160
[imported] - Pseudomonas aeruginosa (strain PAO1),
partial (26%)
Length = 567
Score = 33.9 bits (76), Expect = 0.17
Identities = 29/91 (31%), Positives = 40/91 (43%), Gaps = 17/91 (18%)
Frame = -3
Query: 3 TAISTTTTTKNTKRAPT--------PTRPPLLPSESDNAIVR---------KPKAREVTS 45
T + TTT+ T+ APT P PP +P+ + A R PKAR +S
Sbjct: 505 TTSTMKTTTRTTRTAPTTTCPGTAAPKAPPTIPTSTPCACARCVTSSPPCCSPKARR*SS 326
Query: 46 RYMYSSSSSSSFSSPPRRTNSPLVNRTVNST 76
M S+ S + + P RT R+V ST
Sbjct: 325 PAMNSAVPSMATTMPTART-----ARSVGST 248
>BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 -
Caenorhabditis elegans, partial (1%)
Length = 627
Score = 33.9 bits (76), Expect = 0.17
Identities = 33/106 (31%), Positives = 45/106 (42%)
Frame = +2
Query: 272 NESVTSGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVP 331
N SVTS + S + P SA + + PSS + ST P LVP
Sbjct: 323 NSSVTSAAPSFTAEAETTSPVSSASSTSLETLDATTTAVPSSSIPE--SSSTSLPPTLVP 496
Query: 332 KKSVFDSPASSPRGIVNNRLQGSPIRSAVRPASPSKLGTPSPRSPS 377
+ + SPASSP I + S S+ +S S +P+P PS
Sbjct: 497 QNTT--SPASSPSSITTSTTSSSSSSSSSTTSSSSS-SSPTPPGPS 625
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 33.9 bits (76), Expect = 0.17
Identities = 28/108 (25%), Positives = 49/108 (44%), Gaps = 10/108 (9%)
Frame = +3
Query: 12 KNTKRAPTPTRPPLLPSESDNA---IVRKPKAREVTSRYMYSSSSSSSFSSPPRRT--NS 66
K++ A +PT + P+ S A + + P A + + + SSPP +S
Sbjct: 231 KSSPPASSPTAATVTPAVSPAAPVPVAKSPAASSPVVAPVSTPPKPAPVSSPPAPVPVSS 410
Query: 67 PLVNRTVNSTKQKSTPSVTPAAFV-----KRSQSTERQRQRQGTPRPN 109
P V+S STP+VTP+A V +S+ ++ ++ P P+
Sbjct: 411 PPTPVPVSSPPTASTPAVTPSAEVPAAAPSKSKKKTKKGKKHSAPAPS 554
>BQ751815 similar to PIR|S58652|S586 hypothetical protein YFR036w-a - yeast
(Saccharomyces cerevisiae), partial (9%)
Length = 719
Score = 33.5 bits (75), Expect = 0.23
Identities = 39/125 (31%), Positives = 52/125 (41%), Gaps = 2/125 (1%)
Frame = +1
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T+ S+TT + K P+ TRPP ++S A R P + SSS S S PR
Sbjct: 205 TSFSSTTRPEPCK-PPSSTRPPPKGNKSP-ANYRAPTPNSPRAGTPSSSSPSWSCRPTPR 378
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPR--PNGETPVAQKMLF 120
S + T S +TP P F + ST++ R R +P NG P
Sbjct: 379 SRTSSSPSSTARSAS*STTP--RPRGF--WTTSTDKLRPRSKSPFF*ENGTAPSLPSWSA 546
Query: 121 TSARS 125
T RS
Sbjct: 547 TRRRS 561
>TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophila
melanogaster}, partial (71%)
Length = 698
Score = 33.5 bits (75), Expect = 0.23
Identities = 42/158 (26%), Positives = 64/158 (39%), Gaps = 2/158 (1%)
Frame = +3
Query: 3 TAISTTTTTKNTKRA--PTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSP 60
T STT+ + R+ +P+R +P +A +P + VT R M S S S P
Sbjct: 288 TRSSTTSCPSSRMRS*RSSPSRSRPVPV---SAPASRPSSSSVTPRAM-----SVSVSRP 443
Query: 61 PRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLF 120
PR++ + +PS+ FV + G P TP + L
Sbjct: 444 PRKS-------LLLFALPSLSPSLASFPFVSAT----------GVPTSVSPTPCPPRSLA 572
Query: 121 TSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTP 158
++ S S S + +PVS PPS+ + SL TP
Sbjct: 573 SAVPSPSASSPPPA--VPVSSPPPPSSVSSSSLVSRTP 680
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,503,228
Number of Sequences: 36976
Number of extensions: 223089
Number of successful extensions: 1815
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 1647
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1762
length of query: 573
length of database: 9,014,727
effective HSP length: 101
effective length of query: 472
effective length of database: 5,280,151
effective search space: 2492231272
effective search space used: 2492231272
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140671.7


