

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140670.5 + phase: 0 /pseudo/partial
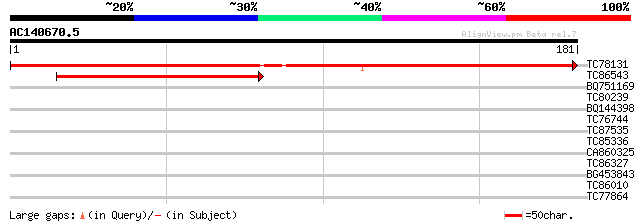
(181 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78131 similar to GP|6091722|gb|AAF03434.1| putative ribosomal ... 291 1e-79
TC86543 similar to GP|4836870|gb|AAD30573.1| 50S Ribosomal prote... 69 8e-13
BQ751169 similar to GP|15021559|gb ORF167 {shrimp white spot syn... 29 0.89
TC80239 similar to GP|7208779|emb|CAB76912.1 hypothetical protei... 28 2.0
BQ144398 similar to GP|13122418|dbj putative glycin-rich protein... 27 3.4
TC76744 similar to SP|Q9FKC0|R1AD_ARATH 60S ribosomal protein L1... 27 4.4
TC87535 similar to GP|19310542|gb|AAL85004.1 unknown protein {Ar... 27 4.4
TC85336 similar to PIR|E86154|E86154 hypothetical protein AAG008... 27 5.8
CA860325 weakly similar to GP|12654479|gb| Unknown (protein for ... 27 5.8
TC86327 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 ... 27 5.8
BG453843 similar to GP|19423912|gb putative 60S ribosomal protei... 26 7.6
TC86010 homologue to PIR|T00795|T00795 26S proteasome regulatory... 26 7.6
TC77864 weakly similar to PIR|T06329|T06329 symbiotic ammonium t... 26 9.9
>TC78131 similar to GP|6091722|gb|AAF03434.1| putative ribosomal protein L13
{Arabidopsis thaliana}, partial (87%)
Length = 953
Score = 291 bits (744), Expect = 1e-79
Identities = 156/191 (81%), Positives = 162/191 (84%), Gaps = 10/191 (5%)
Frame = +2
Query: 1 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 60
RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA
Sbjct: 134 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 313
Query: 61 KDIAVTGRKLTDKVYYWHTGKEL*RIRWPKTLQMLFAKLFCA*FQKTTYVM--------- 111
KDIAVTGRKLTDKVYYWHTG + ++ +TL+ AK +K M
Sbjct: 314 KDIAVTGRKLTDKVYYWHTG-YVGHLK-QRTLKDQMAKDPTDVIRKAILRMIPKNNLRDD 487
Query: 112 -DRKLRIFPGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQN 170
DRKLRIFPGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQN
Sbjct: 488 RDRKLRIFPGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQN 667
Query: 171 AEEMKNSKKSE 181
AEEMKNSKKSE
Sbjct: 668 AEEMKNSKKSE 700
>TC86543 similar to GP|4836870|gb|AAD30573.1| 50S Ribosomal protein L13
{Arabidopsis thaliana}, partial (65%)
Length = 1058
Score = 69.3 bits (168), Expect = 8e-13
Identities = 33/66 (50%), Positives = 46/66 (69%)
Frame = +1
Query: 16 WRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKVY 75
W + DA +ILGRLAS IA + GK+ TYTP+ D G IV+NA+ +AV+G+K T K+Y
Sbjct: 394 WYIVDATDKILGRLASTIANHIRGKNLVTYTPSVDMGSFVIVINAEKVAVSGKKRTQKLY 573
Query: 76 YWHTGK 81
H+G+
Sbjct: 574 RRHSGR 591
>BQ751169 similar to GP|15021559|gb ORF167 {shrimp white spot syndrome
virus}, partial (0%)
Length = 682
Score = 29.3 bits (64), Expect = 0.89
Identities = 15/31 (48%), Positives = 16/31 (51%)
Frame = -2
Query: 123 HPFGDRPLEPYVMPPRTVREMRPRARRAMIR 153
HP D P P +PP R RPR RRA R
Sbjct: 504 HPSDDAPTAP--LPPLPPRHHRPRCRRAPFR 418
>TC80239 similar to GP|7208779|emb|CAB76912.1 hypothetical protein {Cicer
arietinum}, partial (43%)
Length = 930
Score = 28.1 bits (61), Expect = 2.0
Identities = 13/44 (29%), Positives = 25/44 (56%)
Frame = +3
Query: 138 RTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
R R +RA+ +AQK+AEQ+++ E+R + + + +E
Sbjct: 201 RKKRNAEKAQQRALYKAQKEAEQKEKEREKRAKKKGKRKTVTTE 332
>BQ144398 similar to GP|13122418|dbj putative glycin-rich protein {Oryza
sativa (japonica cultivar-group)}, partial (10%)
Length = 1322
Score = 27.3 bits (59), Expect = 3.4
Identities = 15/42 (35%), Positives = 22/42 (51%)
Frame = +3
Query: 139 TVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKS 180
+ R +PR + R +KK E + ++NA KNSKKS
Sbjct: 468 SARRKKPRNEKKSPRREKKTEATPQ*P*IKRNANGGKNSKKS 593
>TC76744 similar to SP|Q9FKC0|R1AD_ARATH 60S ribosomal protein L13a-4.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 911
Score = 26.9 bits (58), Expect = 4.4
Identities = 16/59 (27%), Positives = 30/59 (50%)
Frame = +3
Query: 18 VFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKVYY 76
V DA+ +LGRLAS +A ++ +G +V+ ++I ++G + K+ Y
Sbjct: 90 VIDARHHMLGRLASIVAKELL------------NGQKVVVVRCEEICMSGGLVRQKMKY 230
>TC87535 similar to GP|19310542|gb|AAL85004.1 unknown protein {Arabidopsis
thaliana}, partial (86%)
Length = 893
Score = 26.9 bits (58), Expect = 4.4
Identities = 12/44 (27%), Positives = 25/44 (56%)
Frame = +1
Query: 138 RTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
RT + R + + Q K E++++ ++++ + KNSKKS+
Sbjct: 13 RTKEKKRKENTSSSFQKQSKREKERQK*KKKETKNKEKNSKKSK 144
>TC85336 similar to PIR|E86154|E86154 hypothetical protein AAG00893.1
[imported] - Arabidopsis thaliana, partial (82%)
Length = 1296
Score = 26.6 bits (57), Expect = 5.8
Identities = 14/35 (40%), Positives = 17/35 (48%)
Frame = -3
Query: 89 PKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEH 123
PKT + + +FC F Y D K R PG EH
Sbjct: 811 PKTKPVKYISIFCDFFSVDAYA*DEKCR-KPGMEH 710
>CA860325 weakly similar to GP|12654479|gb| Unknown (protein for
IMAGE:2961467) {Homo sapiens}, partial (10%)
Length = 413
Score = 26.6 bits (57), Expect = 5.8
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = +2
Query: 138 RTVREMRPRARRAMIRAQKKAEQQQENAEQRQN 170
R VRE+ + R +A+KKAE+++ E R+N
Sbjct: 215 RKVREIAEKQRLEEEKARKKAEERRLYLENREN 313
>TC86327 similar to GP|14335106|gb|AAK59832.1 At2g41900/T6D20.20 {Arabidopsis
thaliana}, partial (50%)
Length = 3018
Score = 26.6 bits (57), Expect = 5.8
Identities = 12/43 (27%), Positives = 24/43 (54%)
Frame = +2
Query: 132 PYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEM 174
P M PR++ + P + R AQ++ +QQQ+ + ++ E+
Sbjct: 1976 PGRMSPRSMEALSPMSSRLSAFAQREKQQQQQQQLRSLSSREL 2104
>BG453843 similar to GP|19423912|gb putative 60S ribosomal protein
{Arabidopsis thaliana}, partial (84%)
Length = 608
Score = 26.2 bits (56), Expect = 7.6
Identities = 14/59 (23%), Positives = 28/59 (46%)
Frame = +1
Query: 18 VFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKVYY 76
V DA+G +LGRLAS A ++ G +++ +++ ++G + + Y
Sbjct: 46 VIDAQGHMLGRLASIAAKQLL------------TGQQLVIVRCEEVCISGGLVRQRAKY 186
>TC86010 homologue to PIR|T00795|T00795 26S proteasome regulatory subunit
[imported] - Arabidopsis thaliana, partial (35%)
Length = 1865
Score = 26.2 bits (56), Expect = 7.6
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 5/41 (12%)
Frame = +2
Query: 131 EPYVMPPRTVREMRPRA-----RRAMIRAQKKAEQQQENAE 166
+P +P T P A +A RA KKAE+Q+ NAE
Sbjct: 707 KPTTVPTTTSTVKLPTAVLSTSAKAKARASKKAEEQKANAE 829
>TC77864 weakly similar to PIR|T06329|T06329 symbiotic ammonium transport
protein SAT1 - soybean, partial (21%)
Length = 1411
Score = 25.8 bits (55), Expect = 9.9
Identities = 14/57 (24%), Positives = 24/57 (41%)
Frame = +3
Query: 125 FGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
FG+ +EP + P + R R + A KK + E + AE + + +E
Sbjct: 432 FGNSSIEPIIEPMSHKTKRRTDESRGVKEATKKVRRSCETVQDHLMAERKRRRELTE 602
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,612,369
Number of Sequences: 36976
Number of extensions: 52982
Number of successful extensions: 276
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 274
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 275
length of query: 181
length of database: 9,014,727
effective HSP length: 90
effective length of query: 91
effective length of database: 5,686,887
effective search space: 517506717
effective search space used: 517506717
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC140670.5


