

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140550.12 + phase: 0
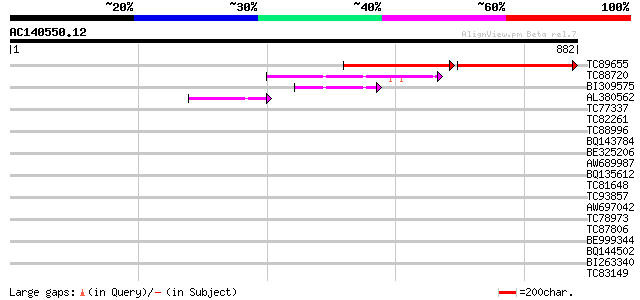
(882 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89655 similar to GP|15408696|dbj|BAB64102. hypothetical protei... 358 0.0
TC88720 similar to GP|15450498|gb|AAK96542.1 AT5g02940/F9G14_250... 97 3e-20
BI309575 similar to GP|15081783|gb AT5g02940/F9G14_250 {Arabidop... 66 7e-11
AL380562 similar to GP|22773250|g Unknown protein {Oryza sativa ... 51 2e-06
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 40 0.003
TC82261 similar to GP|10177249|dbj|BAB10717. contains similarity... 39 0.007
TC88996 similar to PIR|T00933|T00933 RNA-binding protein homolog... 37 0.033
BQ143784 weakly similar to SP|P33485|VNUA Probable nuclear antig... 36 0.056
BE325206 similar to GP|9279699|dbj emb|CAB10253.1~gene_id:MWI23.... 36 0.074
AW689987 weakly similar to GP|1911338|gb|A unknown {Orgyia pseud... 36 0.074
BQ135612 similar to GP|15028978|emb type II keratin E2 {Oncorhyn... 36 0.074
TC81648 similar to PIR|T00840|T00840 probable senescence-related... 35 0.13
TC93857 similar to SP|Q05049|MUC1_XENLA Integumentary mucin C.1 ... 35 0.16
AW697042 similar to GP|13122418|dbj putative glycin-rich protein... 35 0.16
TC78973 similar to PIR|D84745|D84745 plastid protein [imported] ... 34 0.21
TC87806 similar to GP|21592931|gb|AAM64881.1 phosphoserine amino... 34 0.21
BE999344 similar to PIR|T29150|T291 hypothetical protein F47B3.4... 34 0.28
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 34 0.28
BI263340 weakly similar to PIR|T49223|T492 uclacyanin 3 [importe... 34 0.28
TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk p... 33 0.37
>TC89655 similar to GP|15408696|dbj|BAB64102. hypothetical protein~similar
to Arabidopsis thaliana chromosome 1 K9P8.10, partial
(36%)
Length = 1467
Score = 358 bits (918), Expect(2) = 0.0
Identities = 185/186 (99%), Positives = 186/186 (99%)
Frame = +1
Query: 697 ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKS 756
ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSL+LSGFSHNSWIREMQQASDKS
Sbjct: 520 ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLKLSGFSHNSWIREMQQASDKS 699
Query: 757 IIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCI 816
IIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCI
Sbjct: 700 IIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCI 879
Query: 817 KPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFV 876
KPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFV
Sbjct: 880 KPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFV 1059
Query: 877 VLASGE 882
VLASGE
Sbjct: 1060VLASGE 1077
Score = 357 bits (917), Expect(2) = 0.0
Identities = 172/173 (99%), Positives = 173/173 (99%)
Frame = +2
Query: 519 NAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVI 578
NAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVI
Sbjct: 2 NAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVI 181
Query: 579 AEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELW 638
AEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELW
Sbjct: 182 AEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELW 361
Query: 639 MFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSI 691
MFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDS+
Sbjct: 362 MFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSM 520
>TC88720 similar to GP|15450498|gb|AAK96542.1 AT5g02940/F9G14_250
{Arabidopsis thaliana}, partial (35%)
Length = 1773
Score = 97.1 bits (240), Expect = 3e-20
Identities = 83/311 (26%), Positives = 137/311 (43%), Gaps = 37/311 (11%)
Frame = +2
Query: 400 DFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEG 459
D ++ +S + + ++ + +KARAII+L + + D A VL+L + +
Sbjct: 17 DLYHIDLLSKSCTLSLTKSFERAAANKARAIIILPTKGDRFEVDTDAFLTVLALQPIPKM 196
Query: 460 LRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFEN 519
+VE+S L+K + G + V DV +L +QC+ Q GL +I+ +L +
Sbjct: 197 DSVPTIVEVSRSQTCDLLKSISGLKVSPV--EDVASKLFVQCSRQKGLIKIYRHLLNYRK 370
Query: 520 AEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA 579
F + P+L+ + ++ I F DA+ CG+ GKI +P+D+ +L D+VL I
Sbjct: 371 NVFNLCSLPDLEGMTYRQIRHIFKDAVVCGL---YRNGKIYFHPNDDEILXITDKVLFIG 541
Query: 580 EDDDTYAPGPLP------------------------EVRKGYFPRIRDPPKY-------- 607
+T P + E+ K I P
Sbjct: 542 SLXNTNKPQVITTNGKEGKDGINNNELLEKDFDYAIELSKVRLANIVTRPNRSGSKASDG 721
Query: 608 ----PEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKER-ERKLAAGELDVFGL 662
E IL GWR D+ +MI E +L PGS L + +E P +R R+ AG L
Sbjct: 722 NVGPKECILLLGWRPDVVEMIQEYENYLGPGSGLEVLSETPLNDRIIRENIAGHSK---L 892
Query: 663 ENIKLVHREGN 673
+N+++ HR GN
Sbjct: 893 KNVRVSHRIGN 925
Score = 29.3 bits (64), Expect = 6.9
Identities = 12/38 (31%), Positives = 21/38 (54%)
Frame = +3
Query: 488 VVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIK 525
V A +++ + Q A L ++W+DIL + E Y+K
Sbjct: 1197 VAAEEILSLVTAQVAENSELNEVWKDILDADGDEIYVK 1310
>BI309575 similar to GP|15081783|gb AT5g02940/F9G14_250 {Arabidopsis
thaliana}, partial (25%)
Length = 695
Score = 65.9 bits (159), Expect = 7e-11
Identities = 42/136 (30%), Positives = 71/136 (51%)
Frame = +2
Query: 443 DARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCA 502
D A VL+L + + +VE+S L+K + G + V DV +L +QC+
Sbjct: 296 DTDAFLTVLALQPIPKMDSVPTIVEVSRSQTCDLLKSISGLKVSPV--EDVASKLFVQCS 469
Query: 503 LQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVIN 562
Q GL +I+ +L + F + P+L+ + ++ I F DA+ CG+ GKI +
Sbjct: 470 RQKGLIKIYRHLLNYRKNVFNLCSLPDLEGMTYRQIRHIFKDAVVCGL---YRNGKIYFH 640
Query: 563 PDDNYVLRDGDEVLVI 578
P+D+ +L+ D+VLVI
Sbjct: 641 PNDDEILQITDKVLVI 688
>AL380562 similar to GP|22773250|g Unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (11%)
Length = 458
Score = 51.2 bits (121), Expect = 2e-06
Identities = 30/131 (22%), Positives = 66/131 (49%), Gaps = 3/131 (2%)
Frame = +3
Query: 279 SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSL 338
S+ E W +W + + H + +R++ ++ G+L ++ +L +++ + L
Sbjct: 45 SLEECFWEAWACLVSSSTHLKQPTRIERVIGFVLAIWGILFYSRLLSTMTEQFRSNMQRL 224
Query: 339 RKG-KSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEM--DIA 395
R+G + +V+E +H++I G + L +LKQL NK V + A K+ + + D+
Sbjct: 225 REGAQMQVMETDHIIICGMNSHLPFILKQL---NKYHEFAVRLGTASARKQRILLMSDLP 395
Query: 396 KLEFDFMGTSV 406
+ + D + S+
Sbjct: 396 RKQIDRIADSI 428
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 40.4 bits (93), Expect = 0.003
Identities = 34/119 (28%), Positives = 53/119 (43%), Gaps = 5/119 (4%)
Frame = -2
Query: 27 SLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPPPSKPP 86
S +L S TP + + + +S++ + S W+ PS S+S PP PPS
Sbjct: 724 SSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPS--SSSSSSPFSSAPPAPPSGAF 551
Query: 87 VNLIP-----PHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSPIFYLLVICCI 140
L+ P+P P S +D++ + S P PSSSS + S S+ CC+
Sbjct: 550 FELLLLLELLPYPPPSS-DDNSSSPSPNAPSPSSSSSSSSSLPSSPSAS-------CCL 398
>TC82261 similar to GP|10177249|dbj|BAB10717. contains similarity to
phytocyanin/early nodulin-like protein~gene_id:K19P17.3,
partial (7%)
Length = 740
Score = 39.3 bits (90), Expect = 0.007
Identities = 26/99 (26%), Positives = 41/99 (41%), Gaps = 2/99 (2%)
Frame = +1
Query: 56 SEQQWNYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSS 115
SE N PSF + P P S P +I P P LS H+ S+ P PSS
Sbjct: 439 SESPSNSPSFFLPSLLTSPELSPSPSSSPSPSPVISPSPTVLSTPPHSSWISAPSPAPSS 618
Query: 116 SSITKQQQQHSTSSPIFYLLV--ICCIILVPYSAYLQYK 152
+ + Q S + + V ++LV + ++ ++
Sbjct: 619 NQMNSQSSNPKHHSVVIWSTVGGFSFLVLVSFMVFICFR 735
>TC88996 similar to PIR|T00933|T00933 RNA-binding protein homolog At2g42240
- Arabidopsis thaliana, partial (51%)
Length = 651
Score = 37.0 bits (84), Expect = 0.033
Identities = 21/56 (37%), Positives = 27/56 (47%), Gaps = 1/56 (1%)
Frame = +2
Query: 76 RQPPPPPSKPPVNLIPPHP-RPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSP 130
+QPPPPP P+ +IPP P PL H P P I +QQQ + + P
Sbjct: 101 QQPPPPPQYQPIPVIPPPPGAPLPPPHH--------PHPHPQYIPQQQQVYDSYVP 244
>BQ143784 weakly similar to SP|P33485|VNUA Probable nuclear antigen. [strain
Kaplan PRV] {Pseudorabies virus}, partial (2%)
Length = 1022
Score = 36.2 bits (82), Expect = 0.056
Identities = 36/126 (28%), Positives = 50/126 (39%), Gaps = 12/126 (9%)
Frame = +1
Query: 17 PPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGST--SRK 74
P +KT LP+ + S P P G GG + +Q P G G + S+
Sbjct: 97 PAPRKTHHLPATH--PSPRGPRPGGKGGRGG--------APRQIPPPPGPGAGGSQDSQP 246
Query: 75 RRQPPP--PPSK--------PPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQ 124
+ +PPP PP K PP N I PHPRP + PQ ++ K + +
Sbjct: 247 KSKPPPCQPPKKDAPGDTRHPPGNPITPHPRPTRAENGG-------PQEKATGRAKNRPR 405
Query: 125 HSTSSP 130
S P
Sbjct: 406 KKRSPP 423
>BE325206 similar to GP|9279699|dbj emb|CAB10253.1~gene_id:MWI23.18~strong
similarity to unknown protein {Arabidopsis thaliana},
partial (38%)
Length = 662
Score = 35.8 bits (81), Expect = 0.074
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Frame = -2
Query: 78 PPPPPSK----PPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQ 124
P P PS+ PP +L P P S++ H T + P P S + +QQQQ
Sbjct: 493 PTPRPSQHTEPPPTSLQKPQPSQYSLSPHLFQTQTRRPWPQQSQLLRQQQQ 341
Score = 29.6 bits (65), Expect = 5.3
Identities = 18/49 (36%), Positives = 23/49 (46%)
Frame = -2
Query: 82 PSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSP 130
P + V +PP PRP S + TS PQPS S++ Q T P
Sbjct: 523 PQQQDVPFLPPTPRP-SQHTEPPPTSLQKPQPSQYSLSPHLFQTQTRRP 380
>AW689987 weakly similar to GP|1911338|gb|A unknown {Orgyia pseudotsugata
single capsid nuclear polyhedrosis virus}, partial (11%)
Length = 675
Score = 35.8 bits (81), Expect = 0.074
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 5/74 (6%)
Frame = +1
Query: 78 PPPPPSKPPVNLIPPHPRP-----LSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSPIF 132
P P P K P+ PP P S+N++N +LLP +SSI S P+
Sbjct: 226 PTPSPMKIPIYPPPPIPSTNSTSSTSINNNNNNKPNLLPPILTSSIPSPLPFADPSKPLP 405
Query: 133 YLLVICCIILVPYS 146
+LL++ + P+S
Sbjct: 406 FLLLLSPPTISPFS 447
>BQ135612 similar to GP|15028978|emb type II keratin E2 {Oncorhynchus
mykiss}, partial (8%)
Length = 938
Score = 35.8 bits (81), Expect = 0.074
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 3/36 (8%)
Frame = +1
Query: 63 PSFLGIGSTSRKRRQPPP---PPSKPPVNLIPPHPR 95
P F G G +K++ PPP PP PP PP PR
Sbjct: 85 PPFFGGGGGGKKKKNPPPFFFPPPPPPKKKTPPPPR 192
Score = 29.6 bits (65), Expect = 5.3
Identities = 14/35 (40%), Positives = 17/35 (48%), Gaps = 3/35 (8%)
Frame = +3
Query: 63 PSFLGIGSTSRKRRQPPP---PPSKPPVNLIPPHP 94
P F G K+++PPP PP PP PP P
Sbjct: 84 PPFFWGGGGGEKKKKPPPFFFPPPPPPQKKNPPPP 188
>TC81648 similar to PIR|T00840|T00840 probable senescence-related protein
[imported] - Arabidopsis thaliana, partial (36%)
Length = 854
Score = 35.0 bits (79), Expect = 0.13
Identities = 20/61 (32%), Positives = 29/61 (46%), Gaps = 10/61 (16%)
Frame = +2
Query: 78 PPPPPSKPPVNLI----------PPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHST 127
PPPPP+ P+ L+ P P S + N T +LLP S SS++ + + S
Sbjct: 155 PPPPPASTPLRLLHRRKLPNRFLSKSPEPFSTSSINNTALNLLPVTSPSSVSAKVKIPSP 334
Query: 128 S 128
S
Sbjct: 335 S 337
>TC93857 similar to SP|Q05049|MUC1_XENLA Integumentary mucin C.1 (FIM-C.1)
(Fragment). [African clawed frog] {Xenopus laevis},
partial (8%)
Length = 763
Score = 34.7 bits (78), Expect = 0.16
Identities = 35/136 (25%), Positives = 59/136 (42%), Gaps = 6/136 (4%)
Frame = +2
Query: 3 KSNEESSNLNVMNKPPLKKTKTLPSLNLRVSVTP-PNPNDNNGIGGTSTTKTDFSEQQWN 61
K N ++ NV KP T T +VTP P PN +TT T+ + +
Sbjct: 218 KPNPTTTTTNVTPKPKPNPTTTT------TNVTPKPKPNH-------TTTTTNVTPK--- 349
Query: 62 YPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSIT-- 119
P +T+ ++P P P+ N + P P+P +H TT+++ +P + T
Sbjct: 350 -PKPSHTTTTTNVTQKPKPNPTTTTTN-VAPKPKP----NHTTTTTNVTQKPKPNHTTTT 511
Query: 120 ---KQQQQHSTSSPIF 132
Q+ H+T++ F
Sbjct: 512 TNVTQKPNHTTTTTKF 559
>AW697042 similar to GP|13122418|dbj putative glycin-rich protein {Oryza
sativa (japonica cultivar-group)}, partial (11%)
Length = 730
Score = 34.7 bits (78), Expect = 0.16
Identities = 21/58 (36%), Positives = 28/58 (48%)
Frame = -2
Query: 62 YPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSIT 119
YP +TS +PPPPP+ PP +PPHP + + SS P PS + T
Sbjct: 672 YPPLPYSPTTSSHLSKPPPPPTPPP---LPPHP------PTHYSASSQCPAPSPTPPT 526
Score = 30.8 bits (68), Expect = 2.4
Identities = 17/35 (48%), Positives = 22/35 (62%)
Frame = -2
Query: 79 PPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQP 113
PPPP+ PP PP+P PL ++ TTSS L +P
Sbjct: 714 PPPPTPPPRPTPPPYP-PL---PYSPTTSSHLSKP 622
>TC78973 similar to PIR|D84745|D84745 plastid protein [imported] -
Arabidopsis thaliana, partial (69%)
Length = 1583
Score = 34.3 bits (77), Expect = 0.21
Identities = 19/52 (36%), Positives = 23/52 (43%)
Frame = +1
Query: 78 PPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSS 129
PPPPP PP+ PP P +H+ P SSS K HS S+
Sbjct: 703 PPPPPPPPPLPTPPP*P------EHSPNLHLSHPLDSSSQCPKPSSNHSISA 840
>TC87806 similar to GP|21592931|gb|AAM64881.1 phosphoserine aminotransferase
{Arabidopsis thaliana}, partial (58%)
Length = 1010
Score = 34.3 bits (77), Expect = 0.21
Identities = 18/50 (36%), Positives = 26/50 (52%)
Frame = +1
Query: 79 PPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTS 128
PPPP+KP P P+P + +TS L P PS + + + Q ST+
Sbjct: 157 PPPPTKPK-----PSPKPKPIPQIASSTSPLDPPPSLKTSSSKPNQSSTT 291
>BE999344 similar to PIR|T29150|T291 hypothetical protein F47B3.4 -
Caenorhabditis elegans, partial (7%)
Length = 596
Score = 33.9 bits (76), Expect = 0.28
Identities = 17/36 (47%), Positives = 19/36 (52%)
Frame = -3
Query: 64 SFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSV 99
SFL I T + PPPP KPP PP P LS+
Sbjct: 576 SFLYISHTLSHQHSPPPPAKKPPSPPKPPPPSLLSL 469
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 33.9 bits (76), Expect = 0.28
Identities = 20/49 (40%), Positives = 22/49 (44%), Gaps = 2/49 (4%)
Frame = -3
Query: 67 GIGSTSRKRRQPPPPPSKPPVNLIPP--HPRPLSVNDHNKTTSSLLPQP 113
G G R R PP PP PP+ PP PR H T +SL P P
Sbjct: 225 GRGHPPRHRPPPPGPPPPPPLGPRPPPHPPRSGPPRPHPPTRTSLSPPP 79
Score = 32.0 bits (71), Expect = 1.1
Identities = 22/61 (36%), Positives = 26/61 (42%)
Frame = -3
Query: 36 PPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPR 95
PP P+ G+G T+ E W P SR R PPP S PP + PP P
Sbjct: 681 PPPPSSPPGMG----TRPSCQENWWAAPPG---ARPSRPSRYIPPPTSPPPAS--PPAPV 529
Query: 96 P 96
P
Sbjct: 528 P 526
Score = 30.8 bits (68), Expect = 2.4
Identities = 19/59 (32%), Positives = 25/59 (42%)
Frame = -2
Query: 70 STSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTS 128
+ S R PPPP S PP L+PP +SLLP+ S ++ S S
Sbjct: 727 TASLARTPPPPPSSPPPAPLLPP----------RNGNASLLPRKLVGSPARRSSLSSES 581
Score = 29.3 bits (64), Expect = 6.9
Identities = 15/35 (42%), Positives = 17/35 (47%), Gaps = 1/35 (2%)
Frame = -2
Query: 63 PSFLGIGSTSRKRRQPPPPPS-KPPVNLIPPHPRP 96
P+ L + S R R PPPPP P PP P P
Sbjct: 256 PAPLPLASRLRPRASPPPPPPPPGPAASAPPRPPP 152
Score = 28.9 bits (63), Expect = 9.0
Identities = 15/37 (40%), Positives = 17/37 (45%)
Frame = -2
Query: 77 QPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQP 113
+PPPPP PP PP P P + TS QP
Sbjct: 163 RPPPPPPPPPQR--PPPPPPPHTHQPEPPTSPPRRQP 59
>BI263340 weakly similar to PIR|T49223|T492 uclacyanin 3 [imported] -
Arabidopsis thaliana, partial (14%)
Length = 681
Score = 33.9 bits (76), Expect = 0.28
Identities = 25/81 (30%), Positives = 31/81 (37%), Gaps = 1/81 (1%)
Frame = +3
Query: 35 TPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHP 94
TPP+ +T T Q + PS S+ PP P+ PPV L PP P
Sbjct: 255 TPPSTGCGYSPPHDPSTSTPSHNQTPSTPS--NPPSSGGYYNSPPSTPTDPPVTLTPPSP 428
Query: 95 -RPLSVNDHNKTTSSLLPQPS 114
P+ T LP PS
Sbjct: 429 STPIDPGTPTVPTPPFLPSPS 491
>TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk protein
{Argiope trifasciata}, partial (3%)
Length = 1177
Score = 33.5 bits (75), Expect = 0.37
Identities = 21/61 (34%), Positives = 26/61 (42%)
Frame = +3
Query: 70 STSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSS 129
STS RR P PP +P HP+P + S P PSS + T Q +T
Sbjct: 126 STSPSRRPRSKTPLPPPPPPLPQHPKPPQKSPPRMRRSK--PSPSSPAATPLWPQRTTPP 299
Query: 130 P 130
P
Sbjct: 300 P 302
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,057,256
Number of Sequences: 36976
Number of extensions: 412702
Number of successful extensions: 7376
Number of sequences better than 10.0: 186
Number of HSP's better than 10.0 without gapping: 4128
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6182
length of query: 882
length of database: 9,014,727
effective HSP length: 105
effective length of query: 777
effective length of database: 5,132,247
effective search space: 3987755919
effective search space used: 3987755919
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140550.12


