

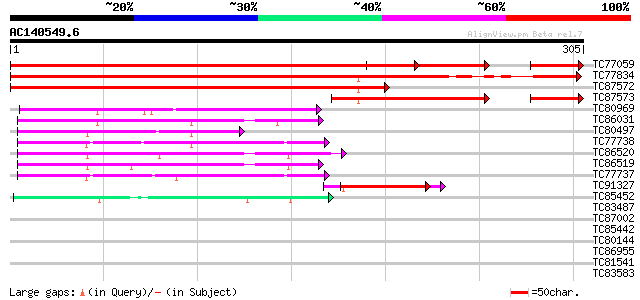
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.6 - phase: 0 /pseudo
(305 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77059 similar to GP|18958499|gb|AAL82617.1 elongation factor 1... 439 e-124
TC77834 similar to GP|18958499|gb|AAL82617.1 elongation factor 1... 342 1e-94
TC87572 similar to GP|18958499|gb|AAL82617.1 elongation factor 1... 341 2e-94
TC87573 similar to GP|18958499|gb|AAL82617.1 elongation factor 1... 151 1e-46
TC80969 similar to GP|1524316|emb|CAA68993.1 glutathione S-trans... 67 9e-12
TC86031 similar to GP|11385459|gb|AAG34812.1 glutathione S-trans... 65 3e-11
TC80497 similar to GP|11385471|gb|AAG34818.1 glutathione S-trans... 64 9e-11
TC77738 homologue to GP|16416392|dbj|BAB70616. glutathione S-tra... 62 3e-10
TC86520 similar to GP|11385463|gb|AAG34814.1 glutathione S-trans... 61 5e-10
TC86519 similar to GP|11385463|gb|AAG34814.1 glutathione S-trans... 61 5e-10
TC77737 similar to GP|16416392|dbj|BAB70616. glutathione S-trans... 57 7e-09
TC91327 similar to GP|22597168|gb|AAN03471.1 unknown protein {Gl... 42 4e-04
TC85452 similar to PIR|T06239|T06239 probable glutathione transf... 40 9e-04
TC83487 40 0.001
TC87002 similar to GP|11385465|gb|AAG34815.1 glutathione S-trans... 40 0.001
TC85442 similar to GP|13543783|gb|AAH06040.1 Unknown (protein fo... 40 0.001
TC80144 similar to PIR|T48205|T48205 hypothetical protein T20L15... 39 0.003
TC86955 similar to GP|11385433|gb|AAG34799.1 glutathione S-trans... 38 0.006
TC81541 similar to GP|22597168|gb|AAN03471.1 unknown protein {Gl... 37 0.009
TC83583 similar to PIR|T06379|T06379 SAR DNA-binding protein 2 -... 36 0.016
>TC77059 similar to GP|18958499|gb|AAL82617.1 elongation factor 1-gamma
{Glycine max}, partial (96%)
Length = 1582
Score = 439 bits (1129), Expect = e-124
Identities = 218/218 (100%), Positives = 218/218 (100%)
Frame = +2
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ
Sbjct: 191 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 370
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV
Sbjct: 371 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 550
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP
Sbjct: 551 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 730
Query: 181 SAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPK 218
SAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPK
Sbjct: 731 SAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPK 844
Score = 94.7 bits (234), Expect(2) = 4e-31
Identities = 45/65 (69%), Positives = 50/65 (76%)
Frame = +3
Query: 191 PKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWKRLYSNTKRNFR 250
PK K +K P+ PK +++ KPKPKNPLDLLPPSKMILDDWKRLYSNTKRNFR
Sbjct: 762 PKPKMNQRKWLNQSPKSPK*KLKRRHRKPKPKNPLDLLPPSKMILDDWKRLYSNTKRNFR 941
Query: 251 EVAIK 255
EVAIK
Sbjct: 942 EVAIK 956
Score = 57.4 bits (137), Expect(2) = 4e-31
Identities = 27/28 (96%), Positives = 28/28 (99%)
Frame = +1
Query: 278 QDSGTCMIQRDTLSGSVITNTMMRTLFL 305
+DSGTCMIQRDTLSGSVITNTMMRTLFL
Sbjct: 955 KDSGTCMIQRDTLSGSVITNTMMRTLFL 1038
>TC77834 similar to GP|18958499|gb|AAL82617.1 elongation factor 1-gamma
{Glycine max}, complete
Length = 1604
Score = 342 bits (877), Expect = 1e-94
Identities = 190/308 (61%), Positives = 221/308 (71%), Gaps = 4/308 (1%)
Frame = +1
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTPQF++MNP+GKVPVLETPDGPVFESNAIARYVAR +NN L+GSSLI AQI+
Sbjct: 220 MGVSNKTPQFLDMNPLGKVPVLETPDGPVFESNAIARYVARQNNNNTLYGSSLIQYAQIE 399
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLE+D+NI K PRLG+ Y PP EE AI+ LKRAL ALN+HLA NTYLVGHS
Sbjct: 400 QWIDFSSLEVDSNINKWLYPRLGYGVYLPPAEEIAITGLKRALNALNTHLASNTYLVGHS 579
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADIITT NLY+GF +L VKSF+SEFPHVERYFWTLVNQPNFRKILGQVKQ EA+ P+
Sbjct: 580 VTLADIITTCNLYVGFTRLFVKSFSSEFPHVERYFWTLVNQPNFRKILGQVKQAEAVQPL 759
Query: 180 PSAKQ--QPKESKPKTKDEPKKVAKSEPEKPKVEVEEEA-PKPKPKNPLDLLPPSKMILD 236
SAK+ QPK+SK K DEPKK AK K + ++ P P P+ L ++
Sbjct: 760 QSAKKPSQPKDSKAKVNDEPKKEAKKGARKAERRSRKKRHPSPNPRIHLIFFLQAR*Y-- 933
Query: 237 DWKRLYSNTKRNFREVAIKDCSKECQNLLTRFNDLIPLFDLQDSGTCMIQRDTLSGSVIT 296
W +T+ +K S + LL++ D GTCMI + TL GSVIT
Sbjct: 934 -WMTGKDSTR------TLKTTS---ERLLSK-----------DFGTCMILKGTLFGSVIT 1050
Query: 297 NTMMRTLF 304
NTMMRTLF
Sbjct: 1051NTMMRTLF 1074
>TC87572 similar to GP|18958499|gb|AAL82617.1 elongation factor 1-gamma
{Glycine max}, partial (55%)
Length = 743
Score = 341 bits (874), Expect = 2e-94
Identities = 170/204 (83%), Positives = 182/204 (88%), Gaps = 2/204 (0%)
Frame = +2
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
MGVSNK+PQFINMNPIGKVPVLETPDGPVFESNA+ARYVARLG +NLFGSS I QA ++Q
Sbjct: 131 MGVSNKSPQFINMNPIGKVPVLETPDGPVFESNAMARYVARLGESNLFGSSPIDQAHVEQ 310
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
WIDFSS+EID NIMKLY PR GF+ Y PVEE AISSLKRA EALN+HLAHNTYLVGHSV
Sbjct: 311 WIDFSSMEIDDNIMKLYRPRRGFSPYLAPVEEAAISSLKRAFEALNTHLAHNTYLVGHSV 490
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADII NLY+GFA+LLVKSFTSEFPHVERYFWTLVNQPNFRKI GQVKQTEAMPPIP
Sbjct: 491 TLADIIAACNLYLGFAQLLVKSFTSEFPHVERYFWTLVNQPNFRKIFGQVKQTEAMPPIP 670
Query: 181 SAKQ--QPKESKPKTKDEPKKVAK 202
SAK QPKE +P +KD PKKVAK
Sbjct: 671 SAKNPTQPKEIQPTSKDAPKKVAK 742
>TC87573 similar to GP|18958499|gb|AAL82617.1 elongation factor 1-gamma
{Glycine max}, partial (50%)
Length = 836
Score = 151 bits (381), Expect(2) = 1e-46
Identities = 76/87 (87%), Positives = 78/87 (89%), Gaps = 3/87 (3%)
Frame = +1
Query: 172 QTEAMPPIPSAKQ--QPKESKPKTKDEPKKVAKSEPEKPKV-EVEEEAPKPKPKNPLDLL 228
QTEAMPPIPS K QPKESKP +KD PKKVAKSEPEKPKV E EEEAPKPKPKNPLDLL
Sbjct: 1 QTEAMPPIPSVKNPTQPKESKPTSKDAPKKVAKSEPEKPKVVEAEEEAPKPKPKNPLDLL 180
Query: 229 PPSKMILDDWKRLYSNTKRNFREVAIK 255
PPSKM+LDDWKRLYSNTK NFREVAIK
Sbjct: 181 PPSKMVLDDWKRLYSNTKTNFREVAIK 261
Score = 52.8 bits (125), Expect(2) = 1e-46
Identities = 25/28 (89%), Positives = 26/28 (92%)
Frame = +2
Query: 278 QDSGTCMIQRDTLSGSVITNTMMRTLFL 305
+DSGTCMIQRD LSGSVITNT MRTLFL
Sbjct: 260 KDSGTCMIQRDILSGSVITNTTMRTLFL 343
>TC80969 similar to GP|1524316|emb|CAA68993.1 glutathione S-transferase
{Petunia x hybrida}, partial (61%)
Length = 811
Score = 67.0 bits (162), Expect = 9e-12
Identities = 48/169 (28%), Positives = 85/169 (49%), Gaps = 8/169 (4%)
Frame = +3
Query: 6 KTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHN---NLFGSSLIHQAQIDQWI 62
K P+F+ + P G+VPV+E D +FES AI RY A + +L G++L +A +DQW+
Sbjct: 186 KKPEFLLLQPFGQVPVVEDGDFRLFESRAIVRYYAAKYADRGPDLLGTTLEEKAVVDQWL 365
Query: 63 DFSSLEID---ANIM--KLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVG 117
+ + D NIM + LP++G + ++ L+ L+ L+ +TYL G
Sbjct: 366 EVEAHNFDDLCFNIMFNLVILPKMGKPGDI-TIAHSSEQKLEEVLDVYERRLSKSTYLAG 542
Query: 118 HSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKI 166
TLAD+ + + SE +V ++ + ++P ++K+
Sbjct: 543 DKFTLADLSHLPGIGHLIEAAKLGHLISERKNVNAWWEKISSRPAWKKL 689
>TC86031 similar to GP|11385459|gb|AAG34812.1 glutathione S-transferase GST
22 {Glycine max}, complete
Length = 1104
Score = 65.1 bits (157), Expect = 3e-11
Identities = 45/175 (25%), Positives = 83/175 (46%), Gaps = 12/175 (6%)
Frame = +3
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHN---NLFGSSLIHQAQIDQW 61
+K P F+ + P G+VP+++ D ++ES AI RY A N +L G ++ + ++QW
Sbjct: 162 HKQPDFLKLQPFGEVPLVQDGDYTLYESRAIIRYYAEKYKNQGTDLLGKTVEERGLVEQW 341
Query: 62 IDFSSLEIDANIMKLYLPRLGFATYFPPVEENAI----SSLKRALEALNSHLAHNTYLVG 117
++ + I L + L P ++ AI LK+ L+ L+ YL G
Sbjct: 342 LEVEAHNFHPAIYSLVINVLFAPLRGVPGDQKAIEESDEKLKKVLDIYEERLSKTKYLAG 521
Query: 118 HSVTLADIITTSNLYIGFAKLLVK-----SFTSEFPHVERYFWTLVNQPNFRKIL 167
+LAD+ ++ F LV + E HV ++ + ++P+++K+L
Sbjct: 522 DFFSLADL-----SHLAFGHYLVNQTGRGNLVRERKHVSAWWDDISSRPSWKKVL 671
>TC80497 similar to GP|11385471|gb|AAG34818.1 glutathione S-transferase GST
10 {Zea mays}, partial (48%)
Length = 675
Score = 63.5 bits (153), Expect = 9e-11
Identities = 41/130 (31%), Positives = 64/130 (48%), Gaps = 9/130 (6%)
Frame = +2
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVA----RLGHNNLFGSSLIHQAQIDQ 60
++ P F+ ++P G+VP + D +FES AI RYV G+ L+G++ + +A IDQ
Sbjct: 137 HRKPDFLKIHPFGQVPAYQDHDISLFESRAICRYVCEKNIEKGNKQLYGTNPLAKASIDQ 316
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFP-PVEENAI----SSLKRALEALNSHLAHNTYL 115
W++ + L +L FA +E AI LK+ L+ + L YL
Sbjct: 317 WLEAEGQSFNPPSSTLVF-QLAFAPRMKIKQDEGAIRQSKEKLKKVLDVYDKRLGETRYL 493
Query: 116 VGHSVTLADI 125
G TLAD+
Sbjct: 494 AGDEFTLADL 523
>TC77738 homologue to GP|16416392|dbj|BAB70616. glutathione S-transferase
{Medicago sativa}, partial (96%)
Length = 748
Score = 62.0 bits (149), Expect = 3e-10
Identities = 51/175 (29%), Positives = 79/175 (45%), Gaps = 9/175 (5%)
Frame = +3
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYV----ARLGHNNLFGSSLIHQAQIDQ 60
+K FI++NP G+VP E D +FES AI +Y+ A G L S A +
Sbjct: 171 HKKEPFISLNPFGQVPAFEDGDLKLFESRAITQYIDHEYADKG-TKLTSSDSKKMAIMSV 347
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFP-PVEENAI----SSLKRALEALNSHLAHNTYL 115
W + SL D + + + LG T F P++ N + + L L+ L+ + YL
Sbjct: 348 WSEVESLHYD-QVASILVWELGIKTLFGIPLDSNVVEENEAKLDIILDVYEKRLSKSKYL 524
Query: 116 VGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQV 170
G S TL D+ +LY K F S P+V + + +P + K+L +
Sbjct: 525 GGDSFTLVDLHHLPSLYYLMKSQSKKLFESR-PYVSAWVADITARPAWSKVLAMI 686
>TC86520 similar to GP|11385463|gb|AAG34814.1 glutathione S-transferase GST
24 {Glycine max}, partial (98%)
Length = 1003
Score = 61.2 bits (147), Expect = 5e-10
Identities = 47/188 (25%), Positives = 86/188 (45%), Gaps = 13/188 (6%)
Frame = +2
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVA---RLGHNNLFGSSLIHQAQIDQW 61
+K P+++ + P G VPV++ D ++ES AI RY A R L G ++ + ++QW
Sbjct: 260 HKDPEYLKLQPFGTVPVIKDGDYTLYESRAIMRYYAEKYRSQGVELLGKTIEEKGLVEQW 439
Query: 62 IDFSSLEIDANIMKLYL-----PRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLV 116
++ + + L LG ++ P V E + + L + L+ N YL
Sbjct: 440 LEVEAHNFHPSAYNLTCHVLCPTLLGGSSPDPKVIEESEAKLVKVFNIYEERLSKNKYLA 619
Query: 117 GHSVTLADIITTSNLYIGFAKLLVKSFTSEF-----PHVERYFWTLVNQPNFRKILGQVK 171
G +LADI ++ F +V + ++ HV ++ + ++P++ K+L K
Sbjct: 620 GDFFSLADI-----SHLPFMDYVVNNMGKDYLIKDKKHVSAWWNDISSRPSWNKVLELYK 784
Query: 172 QTEAMPPI 179
PPI
Sbjct: 785 -----PPI 793
>TC86519 similar to GP|11385463|gb|AAG34814.1 glutathione S-transferase GST
24 {Glycine max}, partial (98%)
Length = 1141
Score = 61.2 bits (147), Expect = 5e-10
Identities = 46/176 (26%), Positives = 84/176 (47%), Gaps = 13/176 (7%)
Frame = +3
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVA---RLGHNNLFGSSLIHQAQIDQW 61
+K P+++ + P G VPV++ D ++ES AI RY A R L G ++ + ++QW
Sbjct: 192 HKDPEYLKLQPFGNVPVIKDGDYTLYESRAIMRYYAEKYRSQGVELLGKTIEEKGLVEQW 371
Query: 62 ID-----FSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLV 116
++ F+ + I L+ L T P V E + + L + L L+ N YL
Sbjct: 372 LEVEAHNFNPPAYNLVIHVLFPSLLVDGTPDPKVIEESEAKLVKVLNIYEERLSKNKYLA 551
Query: 117 GHSVTLADIITTSNLYIGFAKLLVKSFTSEF-----PHVERYFWTLVNQPNFRKIL 167
G +LADI ++ F +V + ++ +V ++ + ++P++ KIL
Sbjct: 552 GDFFSLADI-----SHLPFTDYIVNNMGKDYLIKERKNVSAWWDDISSRPSWNKIL 704
>TC77737 similar to GP|16416392|dbj|BAB70616. glutathione S-transferase
{Medicago sativa}, complete
Length = 911
Score = 57.4 bits (137), Expect = 7e-09
Identities = 50/175 (28%), Positives = 75/175 (42%), Gaps = 9/175 (5%)
Frame = +3
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYV----ARLGHNNLFGSSLIHQAQIDQ 60
+K FI++NP G+VP E D +FES AI +Y+ A G L S A +
Sbjct: 156 HKKEPFISLNPFGQVPAFEDGDLKLFESRAITQYINHEYADKG-TKLTSSDSKKLAIMGV 332
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYF-----PPVEENAISSLKRALEALNSHLAHNTYL 115
W + S D KL + LG T F P + E + L L+ L+ + Y
Sbjct: 333 WSEVESHHFDQVASKL-VWELGIKTLFGIPLDPKIVEENEAKLDSILDVYEKRLSESKYF 509
Query: 116 VGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQV 170
G S TL D+ +L+ K F S PHV + + +P + ++L +
Sbjct: 510 GGDSFTLVDLHHLPSLHYLMKSQSKKLFESR-PHVSAWVADITARPAWSEVLAMI 671
>TC91327 similar to GP|22597168|gb|AAN03471.1 unknown protein {Glycine max},
partial (70%)
Length = 738
Score = 41.6 bits (96), Expect = 4e-04
Identities = 22/50 (44%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Frame = +1
Query: 177 PPIPSAKQQPKE-SKPKTKDEPKKVAK-SEPEKPKVEVEEEAPKPKPKNP 224
P P ++PKE KPK ++PK+ K EPEKPK + P+PKP+ P
Sbjct: 361 PEKPKNPEKPKEPEKPKEPEKPKEPEKPKEPEKPKEKPAPPPPEPKPEPP 510
Score = 40.4 bits (93), Expect = 9e-04
Identities = 28/71 (39%), Positives = 36/71 (50%), Gaps = 6/71 (8%)
Frame = +1
Query: 168 GQVKQTEAM----PPIPSAKQQPKES-KPKTKDEPKKVAK-SEPEKPKVEVEEEAPKPKP 221
G +K E + PP P ++PKE KPK ++PK+ K PEKPK E E+ KP
Sbjct: 250 GAIKSIEIVEPPPPPKPKEPEKPKEPVKPKEPEKPKEPEKPKNPEKPK-EPEKPKEPEKP 426
Query: 222 KNPLDLLPPSK 232
K P P K
Sbjct: 427 KEPEKPKEPEK 459
Score = 38.1 bits (87), Expect = 0.004
Identities = 24/53 (45%), Positives = 29/53 (54%), Gaps = 5/53 (9%)
Frame = +1
Query: 177 PPIPSAKQQPKE-SKPKTKDEPKKVAK-SEPEKPK---VEVEEEAPKPKPKNP 224
P P ++PKE KPK ++PK+ K EPEKPK E E PK KP P
Sbjct: 325 PVKPKEPEKPKEPEKPKNPEKPKEPEKPKEPEKPKEPEKPKEPEKPKEKPAPP 483
Score = 36.6 bits (83), Expect = 0.012
Identities = 21/56 (37%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Frame = +1
Query: 177 PPIPSAKQQPKE-SKPKTKDEPKKVAKSEPEKPKVE--VEEEAPKPKPKNPLDLLP 229
P P ++PKE KPK ++PK+ P +PK E + E PK KP P +P
Sbjct: 397 PEKPKEPEKPKEPEKPKEPEKPKEKPAPPPPEPKPEPPKQPEKPKEKPAPPPQPMP 564
>TC85452 similar to PIR|T06239|T06239 probable glutathione transferase (EC
2.5.1.18) 2 4-D inducible - soybean, partial (92%)
Length = 1115
Score = 40.4 bits (93), Expect = 9e-04
Identities = 46/185 (24%), Positives = 73/185 (38%), Gaps = 15/185 (8%)
Frame = +2
Query: 3 VSNKTPQFINMNPI-GKVPVLETPDGPVFESNAIARYVARLGHNN--LFGSSLIHQAQID 59
+SNK+P + MNP+ K+PVL PV ES +Y+ + ++ S +AQ
Sbjct: 236 LSNKSPLLLQMNPVHKKIPVLIHNGKPVCESLIAVQYIDEVWNDKSPFLPSDPYQKAQAR 415
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
W D+ ID K+Y T V+E A AL+ L L + G
Sbjct: 416 FWADY----ID---KKIYEVAGNLWTKKGEVQETAKKEFIEALKLLEQELGDKNFFGGDK 574
Query: 120 VTLADI--ITTSNLYIGFAKLLVKSFTSEFP----------HVERYFWTLVNQPNFRKIL 167
+ D+ I N + G+ S E P +E +L +Q ++
Sbjct: 575 LGFVDVAFIPLYNWFRGYEAFGKISVDKECPKFFAWANRCMEIESVSKSLPDQDQIHDLI 754
Query: 168 GQVKQ 172
Q+K+
Sbjct: 755 VQIKK 769
>TC83487
Length = 476
Score = 40.0 bits (92), Expect = 0.001
Identities = 34/146 (23%), Positives = 59/146 (40%), Gaps = 9/146 (6%)
Frame = +2
Query: 71 ANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSVTLADIITTSN 130
+NI Y+P P N ++ +KR + LN L YLVG+ TLAD ++
Sbjct: 32 SNIYCRYMPEK-----VPLAINNCVNEVKRVYDVLNKSLEGKEYLVGNRFTLADAVS--- 187
Query: 131 LYIGFAKLLVKSFTSEFPHVERYFWTLVNQ-PNFRKILGQVKQTEAMP--------PIPS 181
+P V +F++ + F + VK+ +A P P+P
Sbjct: 188 ----------------YPIVRGHFFSGIESIDEFSNLKAWVKRIDARPATQKGLNVPVPD 319
Query: 182 AKQQPKESKPKTKDEPKKVAKSEPEK 207
++ +E+ K K+ K + +K
Sbjct: 320 MMKEFRENPEKLKEFEKSIQSFYKDK 397
>TC87002 similar to GP|11385465|gb|AAG34815.1 glutathione S-transferase GST
25 {Glycine max}, partial (94%)
Length = 975
Score = 40.0 bits (92), Expect = 0.001
Identities = 44/168 (26%), Positives = 71/168 (42%), Gaps = 8/168 (4%)
Frame = +3
Query: 8 PQFINMNPIGKVPVLETPDGPVFESNAIARYVA-RLGHNNLFGSSLIHQAQID-QWIDFS 65
P F+ +NP+G VPVL +F+S AI Y+ + + + IH+ I+ Q +
Sbjct: 336 PDFLKLNPVGFVPVLVDGPAVIFDSFAIIMYLEDKFPQQHPLLPTDIHKRAINFQVVSIV 515
Query: 66 SLEI----DANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNT--YLVGHS 119
S I + N +K ++ P V+ S +++ AL L + Y G
Sbjct: 516 SSSIQPLQNLNTLKYIEKKVSPDEKLPWVQ----SVIRKGFTALEKLLKEHAGKYATGDE 683
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKIL 167
V +ADI L + + +EFP + R T P FR+ L
Sbjct: 684 VFMADIFLAPQLDAASKRFNID--MNEFPTLSRLHETYNEIPAFRETL 821
>TC85442 similar to GP|13543783|gb|AAH06040.1 Unknown (protein for MGC:7642)
{Mus musculus}, partial (62%)
Length = 585
Score = 40.0 bits (92), Expect = 0.001
Identities = 18/43 (41%), Positives = 28/43 (64%)
Frame = +3
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPK 222
P K++PK+ +PK ++E K+ AK E EK + E +EE K + K
Sbjct: 63 PEKKEEPKKEEPKKEEEKKEEAKKEEEKKEGEKKEEPKKEEEK 191
>TC80144 similar to PIR|T48205|T48205 hypothetical protein T20L15.110 -
Arabidopsis thaliana, partial (10%)
Length = 682
Score = 38.5 bits (88), Expect = 0.003
Identities = 27/91 (29%), Positives = 39/91 (42%), Gaps = 2/91 (2%)
Frame = +1
Query: 148 PHVERYFWTLVNQPNFRKILGQVKQTEAMPPIPSAKQQPKESKPKTKDEPKKVAKSEPEK 207
P YF +NQ N R PP P K K+ +PK + PK + S +
Sbjct: 295 PRKSYYFTRELNQTNNRIYNTSPPNNTNFPPEPPRKSSSKQQRPKRR-TPKTSSSSPFDS 471
Query: 208 P--KVEVEEEAPKPKPKNPLDLLPPSKMILD 236
P EE P+P+ + LLP + +I+D
Sbjct: 472 PLDSSAESEELPEPEIRTDRVLLPTNDIIID 564
>TC86955 similar to GP|11385433|gb|AAG34799.1 glutathione S-transferase GST
9 {Glycine max}, complete
Length = 1065
Score = 37.7 bits (86), Expect = 0.006
Identities = 23/79 (29%), Positives = 41/79 (51%), Gaps = 3/79 (3%)
Frame = +1
Query: 5 NKTPQFINMNPI-GKVPVLETPDGPVFESNAIARYVARLGHN--NLFGSSLIHQAQIDQW 61
NK+P + MNPI K+PVL + ES I +Y+ + ++ + S +AQ W
Sbjct: 250 NKSPLLLQMNPIHNKIPVLIHNGKSICESAIIVQYIDEVWNDKASFMPSDPYERAQARFW 429
Query: 62 IDFSSLEIDANIMKLYLPR 80
+D+S ++ K++L +
Sbjct: 430 VDYSDKKVYDTWKKMWLSK 486
>TC81541 similar to GP|22597168|gb|AAN03471.1 unknown protein {Glycine max},
partial (36%)
Length = 927
Score = 37.0 bits (84), Expect = 0.009
Identities = 25/66 (37%), Positives = 36/66 (53%), Gaps = 9/66 (13%)
Frame = +3
Query: 168 GQVKQTEAM-PPIPSAKQ--------QPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPK 218
G +K E + PP P A + +PK ++P+ K EP+K K E +KPK +EA K
Sbjct: 516 GSIKSIEIVDPPKPKAAEPEKKKEAEKPKSAEPEKKKEPEK--KKEGDKPK--PAKEAEK 683
Query: 219 PKPKNP 224
PK +P
Sbjct: 684 PKAADP 701
Score = 30.8 bits (68), Expect = 0.68
Identities = 25/83 (30%), Positives = 38/83 (45%), Gaps = 20/83 (24%)
Frame = +3
Query: 171 KQTEAMPPI---PSAKQQPKESK----PKTKDEPKKVAKSEPE-------------KPKV 210
K+ EA P P K++P++ K PK E +K ++PE KPK
Sbjct: 576 KKKEAEKPKSAEPEKKKEPEKKKEGDKPKPAKEAEKPKAADPEKKVTFVSVVKDSDKPK- 752
Query: 211 EVEEEAPKPKPKNPLDLLPPSKM 233
+ E+ PKP+ + P D P+ M
Sbjct: 753 DAEKPKPKPEAEKPKDKPAPTAM 821
>TC83583 similar to PIR|T06379|T06379 SAR DNA-binding protein 2 - garden
pea, partial (33%)
Length = 852
Score = 36.2 bits (82), Expect = 0.016
Identities = 17/47 (36%), Positives = 27/47 (57%)
Frame = +2
Query: 177 PPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKN 223
PP+ K++ KE K K K E K+ + E+P+ EV ++ K K K+
Sbjct: 386 PPVTDKKKEKKEKKEKKKKEKKEEEVEDVEEPEEEVVKKEKKKKKKD 526
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,257,067
Number of Sequences: 36976
Number of extensions: 135567
Number of successful extensions: 1329
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 1057
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1261
length of query: 305
length of database: 9,014,727
effective HSP length: 96
effective length of query: 209
effective length of database: 5,465,031
effective search space: 1142191479
effective search space used: 1142191479
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140549.6


