

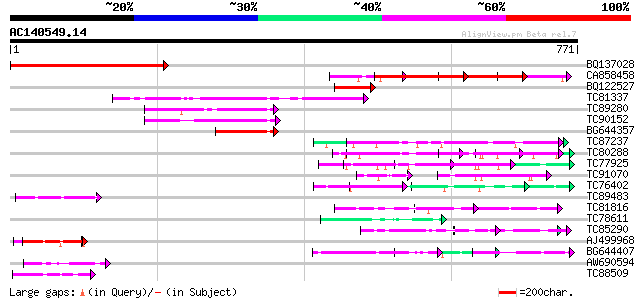
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.14 - phase: 0 /pseudo
(771 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ137028 similar to GP|4914334|gb|A F14N23.20 {Arabidopsis thali... 423 e-118
CA858458 280 2e-75
BQ122527 119 6e-27
TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor... 109 5e-24
TC89280 similar to GP|13278055|gb|AAH03883.1 Similar to RIKEN cD... 100 3e-21
TC90152 homologue to GP|13278055|gb|AAH03883.1 Similar to RIKEN ... 98 1e-20
BG644357 similar to GP|13278055|gb Similar to RIKEN cDNA 2010107... 66 6e-11
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 61 2e-09
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 59 7e-09
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 58 2e-08
TC91070 weakly similar to GP|1086592|gb|AAA82270.1| C. elegans R... 45 2e-06
TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F2... 51 2e-06
TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Ar... 50 2e-06
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 50 4e-06
TC78611 similar to GP|15450599|gb|AAK96571.1 AT5g17440/K3M16_10 ... 49 6e-06
TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 49 6e-06
AJ499968 weakly similar to GP|23495961|gb| hypothetical protein ... 48 2e-05
BG644407 similar to GP|12697612|dbj contains ESTs C97999(C0353) ... 47 4e-05
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 46 5e-05
TC88509 similar to PIR|T01826|T01826 microfibril-associated prot... 45 8e-05
>BQ137028 similar to GP|4914334|gb|A F14N23.20 {Arabidopsis thaliana},
partial (7%)
Length = 687
Score = 423 bits (1088), Expect = e-118
Identities = 203/215 (94%), Positives = 204/215 (94%)
Frame = +2
Query: 1 MWSNRKEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDR 60
MWSNRKEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDR
Sbjct: 41 MWSNRKEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDR 220
Query: 61 IAFEEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIWL 120
IAFEEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIWL
Sbjct: 221 IAFEEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIWL 400
Query: 121 GNEIIIQKKNPNPLLLLHNHNHHHHHHHHQEQHEHHDNRPTSNPFPPESPSQPLHNVAQQ 180
GNEIIIQKKNPNPLLLLHNHNHHHHHHHHQEQHEHHDNRPTSNPFPPESPS AQQ
Sbjct: 401 GNEIIIQKKNPNPLLLLHNHNHHHHHHHHQEQHEHHDNRPTSNPFPPESPSHLCTMFAQQ 580
Query: 181 PPNFGTELDKAHCPFHLKTAACRFGDRCSRVHFYP 215
PPNFG ELDKAHCPFHLKTAACRFGDRC + F P
Sbjct: 581 PPNFGPELDKAHCPFHLKTAACRFGDRC*QGPFLP 685
>CA858458
Length = 386
Score = 280 bits (715), Expect = 2e-75
Identities = 128/128 (100%), Positives = 128/128 (100%)
Frame = +3
Query: 496 SFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRD 555
SFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRD
Sbjct: 3 SFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRD 182
Query: 556 GDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGR 615
GDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGR
Sbjct: 183 GDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGR 362
Query: 616 NSQKWNRD 623
NSQKWNRD
Sbjct: 363 NSQKWNRD 386
Score = 179 bits (454), Expect = 4e-45
Identities = 88/124 (70%), Positives = 100/124 (79%), Gaps = 2/124 (1%)
Frame = +3
Query: 583 SESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDRHR--K 640
++ D R HEEYSDVD T++R+NEK H GRNSQK NRDSR IYEAG DEDR +
Sbjct: 15 NKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGR 194
Query: 641 RHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDNEKQHDSKGKNSQK 700
+HH S+RKGSRH+SRDNSNV DESESDKDRGHE+YSDVDWDTMSR+NEKQH S G+NSQK
Sbjct: 195 KHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQK 374
Query: 701 RNGD 704
N D
Sbjct: 375 WNRD 386
Score = 94.0 bits (232), Expect = 2e-19
Identities = 57/119 (47%), Positives = 72/119 (59%), Gaps = 18/119 (15%)
Frame = +3
Query: 664 SESDKDRGHEKYSDVDWDTMSRDNEKQHDSKGKNSQKRNGDSKYQIDEAGSDEDR--HRK 721
++ D R HE+YSDVD T++RDNEK HD KG+NSQK N DS+ I EAG DEDR +
Sbjct: 15 NKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGR 194
Query: 722 KHLSSQMKESRHQSSDSGYGID--ESDKDKG--------------EMEAQHGYSRKSSR 764
KH S+ K SRHQS D+ D ESDKD+G E E QHG + ++S+
Sbjct: 195 KHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQ 371
Score = 65.5 bits (158), Expect = 7e-11
Identities = 45/124 (36%), Positives = 58/124 (46%), Gaps = 19/124 (15%)
Frame = +3
Query: 436 SSRRRQEDERKQRTLDEEWNTNYKDNHKRKYRNKTSDS---------DSDKEGLEEVDRK 486
+SRR +E R N D R + DS D D++G D +
Sbjct: 27 NSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDG----DGR 194
Query: 487 KYHEHTRKSSFNWNKD----------DNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQK 536
K+H RK S + ++D D R HEEYSDVD T++R+NEK H GRNSQK
Sbjct: 195 KHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQK 374
Query: 537 SNRD 540
NRD
Sbjct: 375 WNRD 386
>BQ122527
Length = 168
Score = 119 bits (297), Expect = 6e-27
Identities = 54/56 (96%), Positives = 56/56 (99%)
Frame = +1
Query: 442 EDERKQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDKEGLEEVDRKKYHEHTRKSSF 497
EDERKQRTLDEEW+TNYKDNHKRKYRN+TSDSDSDKEGLEEVDRKKYHEHTRKSSF
Sbjct: 1 EDERKQRTLDEEWSTNYKDNHKRKYRNETSDSDSDKEGLEEVDRKKYHEHTRKSSF 168
>TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor small
subunit {Arabidopsis thaliana}, partial (81%)
Length = 1190
Score = 109 bits (272), Expect = 5e-24
Identities = 92/347 (26%), Positives = 153/347 (43%)
Frame = +2
Query: 141 NHHHHHHHHQEQHEHHDNRPTSNPFPPESPSQPLHNVAQQPPNFGTELDKAHCPFHLKTA 200
+H +++ H + + H S P ++ L ++ FGTE D+ +CPF+ K
Sbjct: 8 SHRLNNNAHNDSSQFHTLNLRSKP----KMAEHLASI------FGTEKDRVNCPFYFKIG 157
Query: 201 ACRFGDRCSRVHFYPDKSCTLLIKNMYNGPGLAWEQDEGLE*LSGSSPIMA*GAMMQTNG 260
ACR GDRCSR+H P S TLL+ NMY P + G++ PI +
Sbjct: 158 ACRHGDRCSRLHNRPTISPTLLLSNMYQRPDII---TPGVD--PNGQPI---------DP 295
Query: 261 RSLQRV*DCGSKA*KTWMHLKFGEIVNFKVCKNGSFHLRGNVYVQYKLLDSALLAYNSVN 320
R +Q+ + + T + KFG + VC N + H+ GNVYV +K D A A S+
Sbjct: 296 RQIQQHFEDFYEDIFTELS-KFGYVETLNVCDNLADHMIGNVYVLFKEEDHAAAALASLR 472
Query: 321 GRYFAGKQVSCKFVNLTRWKVAICGEYMKSGYKTCSHGTACNFIHCFRNPGGDYEWADSD 380
GR++ G+ + F +T ++ A C +Y ++ +C+ G CNF+H
Sbjct: 473 GRFYEGRPILADFSPVTDFREATCRQYEEN---SCNRGGYCNFMHV-------------- 601
Query: 381 KPPPKFWVKEMVALFGHSDNCEMSRVHGNFSALKHSSNILETDSDREMGQLYGGRSSRRR 440
K + +++V S SR N + S E + R+ GR S R
Sbjct: 602 KKIGRELRRKLV-----SSQRSRSRSRSNSPRRRRRSRSRERERPRDRDYDSRGRRSSDR 766
Query: 441 QEDERKQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDKEGLEEVDRKK 487
R + + ++ +D +R R S+ + +E+ +R+K
Sbjct: 767 DRVRDGDRDRERKRDSRDRDGGRRNGREAREGSEERRARIEQWNREK 907
Score = 36.2 bits (82), Expect = 0.049
Identities = 29/105 (27%), Positives = 47/105 (44%), Gaps = 13/105 (12%)
Frame = +2
Query: 639 RKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTM-------------SR 685
R++ SSQR RSR SN + R E+ D D+D+ R
Sbjct: 623 RRKLVSSQRS----RSRSRSNSPRRRRRSRSRERERPRDRDYDSRGRRSSDRDRVRDGDR 790
Query: 686 DNEKQHDSKGKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMKE 730
D E++ DS+ ++ +RNG + GS+E R R + + + +E
Sbjct: 791 DRERKRDSRDRDGGRRNGREARE----GSEERRARIEQWNREKEE 913
>TC89280 similar to GP|13278055|gb|AAH03883.1 Similar to RIKEN cDNA
2010107D16 gene {Mus musculus}, partial (76%)
Length = 1198
Score = 100 bits (248), Expect = 3e-21
Identities = 62/189 (32%), Positives = 95/189 (49%), Gaps = 7/189 (3%)
Frame = +3
Query: 184 FGTELDKAHCPFHLKTAACRFGDRCSRVHFYPDKSCTLLIKNMYNGPGL-------AWEQ 236
FGTE D+ +CPF+ K ACR GDRCSR+H P S TL++ NMY P + Q
Sbjct: 69 FGTEKDRVNCPFYFKIGACRHGDRCSRLHTKPTISPTLVLSNMYQRPDMNLNFINPTPNQ 248
Query: 237 DEGLE*LSGSSPIMA*GAMMQTNGRSLQRV*DCGSKA*KTWMHLKFGEIVNFKVCKNGSF 296
+ + P +Q + D + K +G+I + +C N +
Sbjct: 249 PQQPQPPQPPQPESLDPDKLQEHFDDFYE--DLFEELSK------YGQIQSLNICDNLAD 404
Query: 297 HLRGNVYVQYKLLDSALLAYNSVNGRYFAGKQVSCKFVNLTRWKVAICGEYMKSGYKTCS 356
H+ GNVYVQYK D A A ++ GR+++G+ + F +T ++ A C +Y ++ C+
Sbjct: 405 HMVGNVYVQYKEEDHAANALMNLTGRFYSGRPIIVDFSPVTDFREATCRQYEEN---VCN 575
Query: 357 HGTACNFIH 365
G CNF+H
Sbjct: 576 RGGYCNFMH 602
>TC90152 homologue to GP|13278055|gb|AAH03883.1 Similar to RIKEN cDNA
2010107D16 gene {Mus musculus}, partial (59%)
Length = 698
Score = 97.8 bits (242), Expect = 1e-20
Identities = 61/188 (32%), Positives = 96/188 (50%), Gaps = 3/188 (1%)
Frame = +2
Query: 184 FGTELDKAHCPFHLKTAACRFGDRCSRVHFYPDKSCTLLIKNMYNGPGLAWEQDEGLE*L 243
FGTE D+ +CPF+ K ACR GDRCSR+H P S T+L+ NMY P +
Sbjct: 146 FGTEKDRVNCPFYFKIGACRHGDRCSRLHTKPSISPTILLSNMYQRPDM----------- 292
Query: 244 SGSSPIMA*GAMMQTNGRSLQRV*DCGSKA*KTWMH--LKFGEIVNFKVCKNGSFHLRGN 301
+ G + +++ D + + K+G+I + VC N + H+ GN
Sbjct: 293 ------ITPGVDVHGQPIDPRKIQDHFEEFYEDLFDELSKYGDIESLNVCDNLADHMVGN 454
Query: 302 VYVQYKLLDSALLAYNSVNGRYFAGKQVSCKF-VNLTRWKVAICGEYMKSGYKTCSHGTA 360
VYVQ++ + A A ++ GR++AG+ + F T ++ A C +Y ++ TC+ G
Sbjct: 455 VYVQFREEEHAGNAVKNLTGRFYAGRPIIVDFSPGGTDFREATCRQYEEN---TCNRGGY 625
Query: 361 CNFIHCFR 368
CNF+H R
Sbjct: 626 CNFMHLKR 649
>BG644357 similar to GP|13278055|gb Similar to RIKEN cDNA 2010107D16 gene
{Mus musculus}, partial (58%)
Length = 576
Score = 65.9 bits (159), Expect = 6e-11
Identities = 30/85 (35%), Positives = 52/85 (60%)
Frame = +2
Query: 281 KFGEIVNFKVCKNGSFHLRGNVYVQYKLLDSALLAYNSVNGRYFAGKQVSCKFVNLTRWK 340
K+GEI + +C N + H+ GNVYVQ++ + A A ++ GR++AG+ + F +T ++
Sbjct: 179 KYGEIESLNICDNLADHMVGNVYVQFREEEQAANALKNLTGRFYAGRPIIVDFSPVTDFR 358
Query: 341 VAICGEYMKSGYKTCSHGTACNFIH 365
A C +Y ++ C+ G CNF+H
Sbjct: 359 EATCRQYEEN---VCNRGGYCNFMH 424
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 60.8 bits (146), Expect = 2e-09
Identities = 74/374 (19%), Positives = 138/374 (36%), Gaps = 28/374 (7%)
Frame = +1
Query: 414 KHSSNILETDSDREMG---QLYGGRSSRRRQEDERKQRTLDEEWNTNYKDNHKRKY---R 467
+ SSN E D++ +L ++S E T + + N + KD + + +
Sbjct: 643 QESSNKTEEQFDKKEKNEFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPK 822
Query: 468 NKTSDSDSDKEGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPH 527
S+SD K+ E+ + K T +S D +++E D N K
Sbjct: 823 GSASESDEQKQEQEQNNTTKDDVQTTDTSSQNGNDTTEKQNETSEDA--------NSKKE 978
Query: 528 DRKGRNSQKSNRDSRDWIYEAGCDE------DRDGDGRKHHISRRKGSRHQSRDNSNVTD 581
D N+ +N DS+ + D DG +H + + NS
Sbjct: 979 DSSALNTTPNNEDSKSGVAGDQADSTTTTSSSETQDGNTNHGEYKDTTNENPEKNSGQEG 1158
Query: 582 ESES--------DKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAG- 632
ES +KD + S E +K S+ + + +++ ++
Sbjct: 1159 TQESGSSSNTFDNKDAASNKVQLTTTSDTSSEQKKDESSSAESKSESSQNDNANSGQSNT 1338
Query: 633 -SDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRD---NE 688
SDE + + S S + + NSN + S+ +++ + D S D NE
Sbjct: 1339 TSDESANDNKDSSQVTTSSENSAEGNSNTENNSDENQNDSKNNENTNDSGNTSNDANVNE 1518
Query: 689 KQHDSKGKNSQKRN-GDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGYGID--ES 745
Q+++ + N GD++ + E+ + + K + + + SSD+ D ES
Sbjct: 1519 NQNENAAQTKTSENEGDAQNESVESKKENNESAHKDVDNNSNSNDQGSSDTSVTQDDKES 1698
Query: 746 DKDKGEMEAQHGYS 759
D G + +G S
Sbjct: 1699 RVDLGTLPESNGES 1740
Score = 60.1 bits (144), Expect = 3e-09
Identities = 58/300 (19%), Positives = 123/300 (40%), Gaps = 5/300 (1%)
Frame = +1
Query: 459 KDNHKRKYRNKTSDSDSDKEGLEEVDRKKYHEHTRKSSF---NWNKDDNSRRHEEYSDVD 515
K + + + +T +S +K LE + K+ E + + S NKD+ + E D
Sbjct: 7 KSHLENEESKETGESSEEKSHLENEESKETGESSEEKSHLENEENKDEEKSKQENEEIKD 186
Query: 516 RVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRD 575
I ++NE+ D + SQ+ N +++ D + +++ + + +G ++
Sbjct: 187 GEKIQQENEENKDEE--KSQQENEENK----------DEEKSQQENELKKNEGG---EKE 321
Query: 576 NSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRN--SQKWNRDSRYRIYEAGS 633
+T+E + +EE S+ + E Q+GS+ + + +DS+ E
Sbjct: 322 TGEITEEKSKQE---NEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSKQGTEETNG 492
Query: 634 DEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDNEKQHDS 693
E ++ H + S + N + E + + E YS + + DN+
Sbjct: 493 TEGGEKEEHDKIKEDTS------SDNQVQDGEKNNEAREENYSGDNASSAVVDNK----- 639
Query: 694 KGKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGYGIDESDKDKGEME 753
SQ+ + ++ Q D+ +E + S++ ES + +ES+KD+ + E
Sbjct: 640 ----SQESSNKTEEQFDKKEKNEFELESQKNSNETTESTDSTITQNSQGNESEKDQAQTE 807
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 58.9 bits (141), Expect = 7e-09
Identities = 52/259 (20%), Positives = 107/259 (41%), Gaps = 18/259 (6%)
Frame = +3
Query: 459 KDNHKRKYRNKTSDSD----SDKEGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDV 514
K NH +K +D+ ++ + + ++ RK H ++ D + HEE +
Sbjct: 261 KHNHDKKKEFDKNDTKLPIRTETDQILKLGRKDLHPGKVEA-------DKNEGHEEEEED 419
Query: 515 DRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSR 574
+ + N N++ HD + + ++ N+ + +E D H R + +++
Sbjct: 420 EHIVYNMQNKREHDEQQQEGEEGNKHETE-------EESED----NVHERREEQDEEENK 566
Query: 575 DNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAGSD 634
+ V +E+ES + +E DV+ D ++EK N R + + + E G D
Sbjct: 567 HGAEVQEENESKSEEVEDEGGDVEID--ENDHEKSEADNDREDEVVDEEKDKE--EEGDD 734
Query: 635 EDRHRKR----------HHSSQRKGSRHRSRDNSNVA---DESESDKDRGHEKYSDVD-W 680
E + + +H + H D+++ A D E+ + G+ ++SD+
Sbjct: 735 ETENEDKEDEEKGGLVENHENHEAREEHYKADDASSAVAHDTHETSTETGNLEHSDLSLQ 914
Query: 681 DTMSRDNEKQHDSKGKNSQ 699
+T +NE H + SQ
Sbjct: 915 NTRKPENETNHSDESYGSQ 971
Score = 50.8 bits (120), Expect = 2e-06
Identities = 29/176 (16%), Positives = 77/176 (43%), Gaps = 6/176 (3%)
Frame = +3
Query: 584 ESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDRHRKRHH 643
E+DK+ GHEE + + + +N+++H + ++ N+ E S+++ H +R
Sbjct: 378 EADKNEGHEEEEEDEHIVYNMQNKREHDEQQQEGEEGNKHET----EEESEDNVHERREE 545
Query: 644 SSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDNEKQHD--SKGKNSQKR 701
+ + N + E + + G + + D + DN+++ + + K+ ++
Sbjct: 546 QDEEENKHGAEVQEENESKSEEVEDEGGDVEIDENDHEKSEADNDREDEVVDEEKDKEEE 725
Query: 702 NGDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGYGI----DESDKDKGEME 753
D D+ ++ + H + + +E +++ D+ + E+ + G +E
Sbjct: 726 GDDETENEDKEDEEKGGLVENHENHEAREEHYKADDASSAVAHDTHETSTETGNLE 893
Score = 45.8 bits (107), Expect = 6e-05
Identities = 30/144 (20%), Positives = 58/144 (39%), Gaps = 9/144 (6%)
Frame = +3
Query: 634 DEDRH------RKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDN 687
+ED H KR H Q++ + N + E +D HE+ + D +
Sbjct: 411 EEDEHIVYNMQNKREHDEQQQEG-----EEGNKHETEEESEDNVHERREEQDEEENKHGA 575
Query: 688 EKQHDSKGKNSQKRNGDSKYQIDE---AGSDEDRHRKKHLSSQMKESRHQSSDSGYGIDE 744
E Q +++ K+ + + +IDE S+ D R+ + + K+ + D D+
Sbjct: 576 EVQEENESKSEEVEDEGGDVEIDENDHEKSEADNDREDEVVDEEKDKEEEGDDETENEDK 755
Query: 745 SDKDKGEMEAQHGYSRKSSRHKRS 768
D++KG + H H ++
Sbjct: 756 EDEEKGGLVENHENHEAREEHYKA 827
Score = 44.7 bits (104), Expect = 1e-04
Identities = 44/187 (23%), Positives = 78/187 (41%), Gaps = 7/187 (3%)
Frame = +3
Query: 439 RRQEDERKQRTLDEEWN---TNYKDNHKRKYRNKTSDSDSDKEGLE-EVDRKKYHEHTRK 494
+R+ DE++Q EE N T + R + D + +K G E + + + E
Sbjct: 447 KREHDEQQQE--GEEGNKHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEEVED 620
Query: 495 SSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDR 554
+ D+N E +D DR D EK + +G + + N D D E++
Sbjct: 621 EGGDVEIDENDHEKSE-ADNDREDEVVDEEKDKEEEG-DDETENEDKED--------EEK 770
Query: 555 DG--DGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVD-WDTMSRENEKQHG 611
G + ++H +R + + ++ D E+ + G+ E+SD+ +T ENE H
Sbjct: 771 GGLVENHENHEAREEHYKADDASSAVAHDTHETSTETGNLEHSDLSLQNTRKPENETNHS 950
Query: 612 SNGRNSQ 618
SQ
Sbjct: 951 DESYGSQ 971
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 57.8 bits (138), Expect = 2e-08
Identities = 62/256 (24%), Positives = 110/256 (42%), Gaps = 23/256 (8%)
Frame = +3
Query: 455 NTNYKDNHKRKYRNKTSDSDSDKEGLEEVDRKKYHEHTRKSSFN---WNKDDNSRRHE-- 509
N +Y D + R S+SD + +GL + ++ + S WN + R E
Sbjct: 288 NGSYLDRNDS--RRNESESDEELKGLSYEEYRRLKRQKMRKSLKHCIWNVTPSPPRREND 461
Query: 510 ---EYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKHHISRR 566
+Y + ++ D +K D++ + KS +S + ++ D + RK
Sbjct: 462 DLEDYGKPEEISEIIDVKK--DKEIKQKAKSESESEE-------SKESDSELRKKRRKSY 614
Query: 567 KGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRY 626
K SR +S S E E K R +YS+ D DT S E +++ ++S++ + SR
Sbjct: 615 KKSR-ESDSESESESEVEDRKRRKSRKYSESDSDTNSEEEDRKRRRRRKSSRRRRKSSRK 791
Query: 627 RIYEAGSDE----------DRHRKRHHSSQRKGSRHRSRDNSNVADE-----SESDKDRG 671
+ + SDE D RKR S + + S+ +S + + E S+S +
Sbjct: 792 KRRCSDSDESETDDESGYDDSGRKRKRSKRSRKSKKKSSEPVSEGSEEIELGSDSAVAKI 971
Query: 672 HEKYSDVDWDTMSRDN 687
+E+ DVD + M+ N
Sbjct: 972 NEEIDDVDEEKMAEMN 1019
Score = 48.9 bits (115), Expect = 7e-06
Identities = 53/260 (20%), Positives = 98/260 (37%), Gaps = 16/260 (6%)
Frame = +3
Query: 524 EKPHDRKGRNSQKSNRDS----------RDWIYEAGCDEDRDGDGRKHHISRRKGSRHQS 573
E P+ R N ++ DS R Y++ D D R+ + RH
Sbjct: 57 ETPYQRHRHNDRRRTPDSDVSNDRPRYRRSPSYDSYDDRHTDRHRRRSISPENRNPRHHD 236
Query: 574 RDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEA-- 631
NSN + R + Y D + D+ E+E G + +++ R R ++ ++
Sbjct: 237 NGNSNGNHLPKKFGHRNNGSYLDRN-DSRRNESESDEELKGLSYEEYRRLKRQKMRKSLK 413
Query: 632 ----GSDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDN 687
R+ + + G + +V + E + E S+ S+++
Sbjct: 414 HCIWNVTPSPPRRENDDLEDYGKPEEISEIIDVKKDKEIKQKAKSESESE-----ESKES 578
Query: 688 EKQHDSKGKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGYGIDESDK 747
+ + K + S K++ +S + + EDR R+ K ++ SDS +E D+
Sbjct: 579 DSELRKKRRKSYKKSRESDSESESESEVEDRKRR-------KSRKYSESDSDTNSEEEDR 737
Query: 748 DKGEMEAQHGYSRKSSRHKR 767
+ RKSSR KR
Sbjct: 738 KRRRRRKSSRRRRKSSRKKR 797
Score = 47.8 bits (112), Expect = 2e-05
Identities = 45/191 (23%), Positives = 78/191 (40%), Gaps = 5/191 (2%)
Frame = +3
Query: 421 ETDSDREMGQLYGGRSSRRRQEDERKQRTLDEEWNT-----NYKDNHKRKYRNKTSDSDS 475
+++S+ E + ++R++ +K R D E + + K RKY SD++S
Sbjct: 543 KSESESEESKESDSELRKKRRKSYKKSRESDSESESESEVEDRKRRKSRKYSESDSDTNS 722
Query: 476 DKEGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQ 535
++E + R+K RKS SR+ SD D D+E +D GR +
Sbjct: 723 EEEDRKRRRRRKSSRRRRKS---------SRKKRRCSDSDE--SETDDESGYDDSGRKRK 869
Query: 536 KSNRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYS 595
+S R + S++K S S + + S+S + +EE
Sbjct: 870 RSKRSRK---------------------SKKKSSEPVSEGSEEIELGSDSAVAKINEEID 986
Query: 596 DVDWDTMSREN 606
DVD + M+ N
Sbjct: 987 DVDEEKMAEMN 1019
Score = 31.6 bits (70), Expect = 1.2
Identities = 25/111 (22%), Positives = 47/111 (41%)
Frame = +3
Query: 4 NRKEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAF 63
+ E+ K RK+RRK + + E+ E+RKR ++ + E D
Sbjct: 552 SESEESKESDSELRKKRRKSYKKSRESDSESESESEVEDRKR----RKSRKYSESDSDTN 719
Query: 64 EEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGR 114
E E +++++R +++ S R + E E +++ DD GR
Sbjct: 720 SEEED----RKRRRRRKSSRRRRKSSRKKRRCSDSDESETDDESGYDDSGR 860
>TC91070 weakly similar to GP|1086592|gb|AAA82270.1| C. elegans RSP-6
protein (corresponding sequence C33H5.12a)
{Caenorhabditis elegans}, partial (17%)
Length = 859
Score = 44.7 bits (104), Expect(2) = 2e-06
Identities = 42/178 (23%), Positives = 75/178 (41%), Gaps = 22/178 (12%)
Frame = +1
Query: 582 ESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEA----GSDEDR 637
E + +DR D DW ++E + + R+ K RD RYR + G D+
Sbjct: 247 ERDMSRDRTRGGERDRDW-----KSEWDYRNRERDINKERRDDRYRHNDEDTPNGKDKHM 411
Query: 638 HRK---------RHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDNE 688
HR+ + HS + H R N E + K +G + DVD DT+ ++
Sbjct: 412 HREDDNDIGDRYKKHSRHEENGYHADRKR-NYTMEEDGRKLKGEVEQDDVDEDTLELPDQ 588
Query: 689 KQHD-SKGKNSQKRNGDS---KY-----QIDEAGSDEDRHRKKHLSSQMKESRHQSSD 737
++ D + K +R ++ KY QI +A +E + + + + + ++ SD
Sbjct: 589 EEEDLIRIKEESRRRREAIMEKYKKQSQQIQQAAENEGKDKDMEIHTDISKAIDGKSD 762
Score = 36.6 bits (83), Expect = 0.037
Identities = 31/132 (23%), Positives = 53/132 (39%)
Frame = +1
Query: 478 EGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKS 537
EG E D + + +W + + R E + +R RD+ H+ + + K
Sbjct: 235 EGKPERDMSRDRTRGGERDRDWKSEWDYRNRERDINKER----RDDRYRHNDEDTPNGKD 402
Query: 538 NRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDV 597
R+ D++ GD K H + H R N T E + K +G E DV
Sbjct: 403 KHMHRE-------DDNDIGDRYKKHSRHEENGYHADRKR-NYTMEEDGRKLKGEVEQDDV 558
Query: 598 DWDTMSRENEKQ 609
D DT+ ++++
Sbjct: 559 DEDTLELPDQEE 594
Score = 25.8 bits (55), Expect(2) = 2e-06
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Frame = +3
Query: 472 DSDSDKEGLEEVDRKKYHEHTRKSSFNW---NKDDNSRRHEEYSDVDRVTINRDNEKPHD 528
D D D++ E KK E +R +W + + R H+ + D V +R E+
Sbjct: 6 DRDVDRDLHRE---KKREETSRNKEVDWVHRREKERGRSHDRHRR-DMVK-DRSRERDEG 170
Query: 529 RKGRNSQKSNRDSRDWIYE 547
R R + +R SRD +YE
Sbjct: 171 RDRRREMERDR-SRDAVYE 224
>TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F23H14.7
{Arabidopsis thaliana}, partial (43%)
Length = 1449
Score = 50.8 bits (120), Expect = 2e-06
Identities = 45/231 (19%), Positives = 81/231 (34%), Gaps = 9/231 (3%)
Frame = +3
Query: 547 EAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRG---------HEEYSDV 597
EA D + ++ + K ++H+ R S+ + +SD +R H ++S +
Sbjct: 756 EAAAKAKADAEAKEKKL---KTAKHKKRSGSDTDSDRDSDDERKRASKRSHRKHRKHSHI 926
Query: 598 DWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDN 657
D + EK S KW R + D SS + R + +
Sbjct: 927 DSGDHEKRKEK--------SSKWKTKKR-----SSESSDFSSDESESSSEEEKRRKKKQR 1067
Query: 658 SNVADESESDKDRGHEKYSDVDWDTMSRDNEKQHDSKGKNSQKRNGDSKYQIDEAGSDED 717
+ D+ G + Y+D +K K +R SK+ + SDE
Sbjct: 1068KKIRDQDSRSNSSGSDSYAD-------------EVNKRKRRHRRRRPSKFSDSDFYSDEG 1208
Query: 718 RHRKKHLSSQMKESRHQSSDSGYGIDESDKDKGEMEAQHGYSRKSSRHKRS 768
+ RH+ S+S D S + ++ + + R H+RS
Sbjct: 1209ECSDRKRGHGKHHKRHRHSESSES-DLSSDESDDIHRKRSHRRHHKHHRRS 1358
Score = 48.1 bits (113), Expect = 1e-05
Identities = 54/250 (21%), Positives = 92/250 (36%), Gaps = 4/250 (1%)
Frame = +3
Query: 462 HKRKYRNKTSDSDSDKEGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINR 521
HK++ SD+DSD++ +E R H R+H ++S +D +
Sbjct: 819 HKKR---SGSDTDSDRDSDDERKRASKRSH--------------RKHRKHSHID----SG 935
Query: 522 DNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTD 581
D+EK RK ++S+ W + E D + S + R + + + D
Sbjct: 936 DHEK---RKEKSSK--------WKTKKRSSESSDFSSDESESSSEEEKRRKKKQRKKIRD 1082
Query: 582 ESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDR---- 637
+ G + Y+D N+++ R K++ Y SD R
Sbjct: 1083 QDSRSNSSGSDSYAD-------EVNKRKRRHRRRRPSKFSDSDFYSDEGECSDRKRGHGK 1241
Query: 638 HRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDNEKQHDSKGKN 697
H KRH S+ S+ +D S + D H K S +H + N
Sbjct: 1242 HHKRHRHSE-----------SSESDLSSDESDDIHRK--------RSHRRHHKHHRRSHN 1364
Query: 698 SQKRNGDSKY 707
++ R+ DS Y
Sbjct: 1365 TEVRSSDSDY 1394
Score = 42.4 bits (98), Expect = 7e-04
Identities = 35/139 (25%), Positives = 59/139 (42%), Gaps = 11/139 (7%)
Frame = +3
Query: 414 KHSSNILETDSDREMGQLYGGRSSRRRQEDERKQRTLDEEWNTNYKDN-----------H 462
K SS + SD RR+++ +K R D N++ D+ H
Sbjct: 981 KRSSESSDFSSDESESS--SEEEKRRKKKQRKKIRDQDSRSNSSGSDSYADEVNKRKRRH 1154
Query: 463 KRKYRNKTSDSDSDKEGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRD 522
+R+ +K SDSD + E DRK+ H K + ++ +E D+ R +R
Sbjct: 1155 RRRRPSKFSDSDFYSDEGECSDRKRGHGKHHKRHRHSESSESDLSSDESDDIHRKRSHRR 1334
Query: 523 NEKPHDRKGRNSQKSNRDS 541
+ K H R+ N++ + DS
Sbjct: 1335 HHK-HHRRSHNTEVRSSDS 1388
>TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1711
Score = 50.4 bits (119), Expect = 2e-06
Identities = 35/117 (29%), Positives = 62/117 (52%), Gaps = 1/117 (0%)
Frame = +1
Query: 9 RKALKKLKRKERRKEIA-EKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEERE 67
R+ +K + ++R +E+ EKER+ E + E ER RI E+++ RIE FE +
Sbjct: 118 RRHDRKSRERDRERELKREKERVLERY---EREAERDRIRKEREQKRRIEEVERQFELQL 288
Query: 68 KAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIWLGNEI 124
K W + ++R+ E+E+Q +++ ++ +E ++ DDD D R W N I
Sbjct: 289 KEW---EYREREKEKERQYEKEKEKDRERKRRKEILYDEEDDDGDSRKR--WRXNAI 444
Score = 37.7 bits (86), Expect = 0.017
Identities = 44/197 (22%), Positives = 85/197 (42%), Gaps = 12/197 (6%)
Frame = +1
Query: 435 RSSRRRQEDERKQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDKEG---LEEVDRKKYHEH 491
R R RQ ++K + + T + H RK R + + + +E LE +R+ +
Sbjct: 46 RKMRERQTGDKKPTSEHDRPETPDR-RHDRKSRERDRERELKREKERVLERYEREAERDR 222
Query: 492 TRKSSFNWNKDDNSRRHEEYSDVDRVTIN----RDNEKPHDRKGRNSQKSNRDSR---DW 544
RK + RR EE + + R+ EK +R+ ++ +R+ + +
Sbjct: 223 IRKER------EQKRRIEEVERQFELQLKEWEYREREKEKERQYEKEKEKDRERKRRKEI 384
Query: 545 IYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESE--SDKDRGHEEYSDVDWDTM 602
+Y+ +ED DGD RK R + + + + N + ++ + +DK + EE + +
Sbjct: 385 LYD---EEDDDGDSRK----RWRXNAIEEKRNKRLREKEDDLADKQKEEEEIA----EAK 531
Query: 603 SRENEKQHGSNGRNSQK 619
R E Q R++ K
Sbjct: 532 KRTEEDQQLKRQRDALK 582
Score = 31.2 bits (69), Expect = 1.6
Identities = 22/85 (25%), Positives = 41/85 (47%), Gaps = 1/85 (1%)
Frame = +2
Query: 411 SALKHSSNILETDSDREMGQLYGGRSSRRRQEDERKQRTLDEEWNTNYKDNHKRKYRNKT 470
+A + + I T+ E RS ++ + R +E TN++D H R +
Sbjct: 983 AAAEFAKRISSTNFKEERLDGERDRSRHSNEKSNHRDRDRSDEDGTNHRDEH----RERN 1150
Query: 471 SDSDSDKE-GLEEVDRKKYHEHTRK 494
SD D D++ GLE + K ++++R+
Sbjct: 1151 SDRDRDRDHGLE---KHKTYDNSRR 1216
Score = 30.8 bits (68), Expect = 2.0
Identities = 43/200 (21%), Positives = 75/200 (37%), Gaps = 2/200 (1%)
Frame = +1
Query: 563 ISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNR 622
I R + + R+ + S+ DR E D D SRE +++ + R
Sbjct: 22 IQRMNRVKRKMRERQTGDKKPTSEHDR--PETPDRRHDRKSRERDRERELKREKERVLER 195
Query: 623 DSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDT 682
YE ++ DR RK +R +E E + + +W+
Sbjct: 196 ------YEREAERDRIRKEREQKRR-------------IEEVERQFELQLK-----EWEY 303
Query: 683 MSRDNEK--QHDSKGKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGY 740
R+ EK Q++ + + ++R + DE D D RK+ + ++E R++
Sbjct: 304 REREKEKERQYEKEKEKDRERKRRKEILYDEEDDDGD-SRKRWRXNAIEEKRNKRLREKE 480
Query: 741 GIDESDKDKGEMEAQHGYSR 760
D +DK K E E R
Sbjct: 481 D-DLADKQKEEEEIAEAKKR 537
Score = 30.0 bits (66), Expect = 3.5
Identities = 22/69 (31%), Positives = 30/69 (42%)
Frame = +2
Query: 509 EEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKHHISRRKG 568
EE D +R NEK + R S + + RD E D DRD R H +
Sbjct: 1028 EERLDGERDRSRHSNEKSNHRDRDRSDEDGTNHRDEHRERNSDRDRD---RDHGL----- 1183
Query: 569 SRHQSRDNS 577
+H++ DNS
Sbjct: 1184 EKHKTYDNS 1210
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 49.7 bits (117), Expect = 4e-06
Identities = 44/202 (21%), Positives = 84/202 (40%), Gaps = 1/202 (0%)
Frame = +3
Query: 551 DEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEKQH 610
D+ GD ++ + K +S + DE + KD+ ++ +DVD ++ EK+
Sbjct: 96 DDKSAGDVKEDKVEIEKDLEIKSVEKD---DEKKEKKDKEKKDKTDVDEGKDKKDKEKKK 266
Query: 611 GSNGRNSQKWNRDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSR-DNSNVADESESDKD 669
+ K G +ED K+ ++K + + + D +E +S KD
Sbjct: 267 KEKKEENVK------------GEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKD 410
Query: 670 RGHEKYSDVDWDTMSRDNEKQHDSKGKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMK 729
+ +K + D D D K+ +K K +K+ D K + + D D + + + K
Sbjct: 411 KEKKKDKNED-DDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDID---IEETAKEGK 578
Query: 730 ESRHQSSDSGYGIDESDKDKGE 751
E + + D +S KDK +
Sbjct: 579 EKKKKKEDKEEKKKKSGKDKDQ 644
Score = 43.9 bits (102), Expect = 2e-04
Identities = 40/209 (19%), Positives = 86/209 (41%), Gaps = 13/209 (6%)
Frame = +3
Query: 442 EDERKQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDKEGLEEVDRKKYHEHTRKSSFNWNK 501
++++ + D E + KD+ K++ ++K DK ++E KK E +K K
Sbjct: 120 KEDKVEIEKDLEIKSVEKDDEKKEKKDKEK---KDKTDVDEGKDKKDKEKKKKEK----K 278
Query: 502 DDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKH 561
++N + EE D ++ D++ + +K + D +D+DG+ +K
Sbjct: 279 EENVKGEEE-----------DGDEKKDKEKKKKEKKEKGKED--------KDKDGEEKKS 401
Query: 562 HISRRK-------------GSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENEK 608
+ K GS+ + + + E +K+ G D+D + ++E
Sbjct: 402 KKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETAKE--- 572
Query: 609 QHGSNGRNSQKWNRDSRYRIYEAGSDEDR 637
G+ +K D + ++G D+D+
Sbjct: 573 -----GKEKKKKKEDKEEKKKKSGKDKDQ 644
Score = 39.7 bits (91), Expect = 0.004
Identities = 25/108 (23%), Positives = 64/108 (59%), Gaps = 1/108 (0%)
Frame = +3
Query: 7 EKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEER 66
EK +K +++ + +KE +KE+ ++ D +E + + E+++ E+ E + EE
Sbjct: 141 EKDLEIKSVEKDDEKKEKKDKEKKDK----TDVDEGKDKKDKEKKKKEKKEENVKGEEED 308
Query: 67 EKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEED-VDDDDDG 113
K++++++ +E+ ++ +D E+ +S+ ++E+++D +DDD+G
Sbjct: 309 GDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDDDEG 452
>TC78611 similar to GP|15450599|gb|AAK96571.1 AT5g17440/K3M16_10 {Arabidopsis
thaliana}, partial (91%)
Length = 1636
Score = 49.3 bits (116), Expect = 6e-06
Identities = 45/172 (26%), Positives = 66/172 (38%)
Frame = +1
Query: 423 DSDREMGQLYGGRSSRRRQEDERKQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDKEGLEE 482
DSDR + +GG+ + K L EE N + K + R+K D D+E +
Sbjct: 904 DSDRRLADHFGGKLHLGYMQIREKLAELQEERNKSRKIDRLDDRRSKERSRDRDREPSRD 1083
Query: 483 VDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQKSNRDSR 542
+R + HE R DN RR + RDN + HDR
Sbjct: 1084 RERGESHERGR---------DNDRRSRD----------RDN-RHHDR------------- 1164
Query: 543 DWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEY 594
+ G D DRD D ++ R+ SR + R + D+ R H+ Y
Sbjct: 1165 ----DRGYDRDRDRDSSRYDSRSRRRSRSRERSR-------DYDRHRRHDRY 1287
Score = 39.3 bits (90), Expect = 0.006
Identities = 30/107 (28%), Positives = 47/107 (43%), Gaps = 10/107 (9%)
Frame = +1
Query: 625 RYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRG--HEKYSDVDWDT 682
R ++ E + ++ RK R+ S+ RSRD E D++RG HE+ D D +
Sbjct: 967 REKLAELQEERNKSRKIDRLDDRR-SKERSRDRDR---EPSRDRERGESHERGRDNDRRS 1134
Query: 683 MSRDNEKQHDSKGKNSQKRNGDSKY--------QIDEAGSDEDRHRK 721
RDN +G + + S+Y + E D DRHR+
Sbjct: 1135 RDRDNRHHDRDRGYDRDRDRDSSRYDSRSRRRSRSRERSRDYDRHRR 1275
Score = 34.3 bits (77), Expect = 0.19
Identities = 28/108 (25%), Positives = 45/108 (40%), Gaps = 3/108 (2%)
Frame = +1
Query: 548 AGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSDVDWDTMSRENE 607
A E+R+ + + R+ S+ +SRD E D++RG D D SR+ +
Sbjct: 979 AELQEERNKSRKIDRLDDRR-SKERSRDRDR---EPSRDRERGESHERGRDNDRRSRDRD 1146
Query: 608 KQHGSNGRNSQKWNRD---SRYRIYEAGSDEDRHRKRHHSSQRKGSRH 652
+H R + +RD SRY R R R + R+ R+
Sbjct: 1147 NRHHDRDRGYDR-DRDRDSSRYDSRSRRRSRSRERSRDYDRHRRHDRY 1287
>TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (32%)
Length = 1558
Score = 49.3 bits (116), Expect = 6e-06
Identities = 33/160 (20%), Positives = 65/160 (40%)
Frame = +3
Query: 605 ENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADES 664
E+ +++ S G S Y Y +++ RKR +K ++ ++
Sbjct: 642 EDTQEYISIGNESYSKEIFEEYITYLKEKAKEKERKREEEKAKKEKEREEKEKRKEKEKK 821
Query: 665 ESDKDRGHEKYSDVDWDTMSRDNEKQHDSKGKNSQKRNGDSKYQIDEAGSDEDRHRKKHL 724
E +++R EK S++ K+ +S N +G + + D++R ++
Sbjct: 822 EKEREREKEK---------SKERHKKDESDSDNQDMTDGHGYREEKKKEKDKERKHRR-- 968
Query: 725 SSQMKESRHQSSDSGYGIDESDKDKGEMEAQHGYSRKSSR 764
RHQSS ++ +K++ +HG RK SR
Sbjct: 969 -------RHQSSMDDVDSEKDEKEESRKSRRHGSDRKKSR 1067
Score = 47.8 bits (112), Expect = 2e-05
Identities = 47/191 (24%), Positives = 78/191 (40%)
Frame = +3
Query: 477 KEGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNSQK 536
KE +E +RK+ E +K +++ +R E+ ++ R+ EK ++ +
Sbjct: 720 KEKAKEKERKREEEKAKKEK---EREEKEKRKEK----EKKEKEREREKEKSKERHKKDE 878
Query: 537 SNRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKDRGHEEYSD 596
S+ D++D G E++ + K RR RHQS D+ +S+KD E
Sbjct: 879 SDSDNQDMTDGHGYREEKKKEKDKERKHRR---RHQSS-----MDDVDSEKDEKEES--- 1025
Query: 597 VDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRD 656
++HGS+ + S+K SD + KRH GSR RS
Sbjct: 1026--------RKSRRHGSDRKKSRKHANSPE-------SDNESRHKRHKRDHGDGSR-RSGW 1157
Query: 657 NSNVADESESD 667
N + D D
Sbjct: 1158NEELEDGELGD 1190
Score = 45.4 bits (106), Expect = 8e-05
Identities = 31/150 (20%), Positives = 60/150 (39%), Gaps = 2/150 (1%)
Frame = +3
Query: 604 RENEKQHGSNGRNSQKWNRDSRYRIYEAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADE 663
RE EK R ++ ++ + E ++++ ++RH + D +E
Sbjct: 750 REEEKAKKEKEREEKEKRKEKEKKEKEREREKEKSKERHKKDESDSDNQDMTDGHGYREE 929
Query: 664 --SESDKDRGHEKYSDVDWDTMSRDNEKQHDSKGKNSQKRNGDSKYQIDEAGSDEDRHRK 721
E DK+R H + D + D+EK + + S++ D K A S E +
Sbjct: 930 KKKEKDKERKHRRRHQSSMDDV--DSEKDEKEESRKSRRHGSDRKKSRKHANSPESDNES 1103
Query: 722 KHLSSQMKESRHQSSDSGYGIDESDKDKGE 751
+H ++ S SG+ + D + G+
Sbjct: 1104RH-KRHKRDHGDGSRRSGWNEELEDGELGD 1190
Score = 40.4 bits (93), Expect = 0.003
Identities = 32/115 (27%), Positives = 61/115 (52%), Gaps = 10/115 (8%)
Frame = +1
Query: 5 RKEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERK-----RILLEQQEAERIERD 59
+KEKRK K +RK+RRKE K+ + + R + K ++ +++ +RI+R+
Sbjct: 778 KKEKRKKNVKRRRKKRRKENVRKKNQKNDIRKMNQIVIIKT*LTAMVIERRRKKKRIKRE 957
Query: 60 RIAFEEREKA----WIIKQQQQRDLE-EEQQQLSQRDLEQHESELEEEEEEDVDD 109
I EE +A WI+K+ +++ LE E +++ R L ++ L + + + D
Sbjct: 958 NI--EEGIRAVWMMWILKKMRRKSLENREDMEVTARSLGSMQTLLSQTMKVGIKD 1116
Score = 39.3 bits (90), Expect = 0.006
Identities = 26/115 (22%), Positives = 55/115 (47%), Gaps = 1/115 (0%)
Frame = +3
Query: 421 ETDSD-REMGQLYGGRSSRRRQEDERKQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDKEG 479
E+DSD ++M +G R +++++D+ ++ H+R++++ D DS+K+
Sbjct: 876 ESDSDNQDMTDGHGYREEKKKEKDKERK--------------HRRRHQSSMDDVDSEKDE 1013
Query: 480 LEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGRNS 534
EE + + H RK S R+H + D + ++ +++ H R S
Sbjct: 1014KEESRKSRRHGSDRKKS---------RKHANSPESDNESRHKRHKRDHGDGSRRS 1151
Score = 38.9 bits (89), Expect = 0.008
Identities = 16/73 (21%), Positives = 36/73 (48%)
Frame = +3
Query: 694 KGKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGYGIDESDKDKGEME 753
K K +++ + K + ++ ++++ ++K + +E + S + DESD D +M
Sbjct: 726 KAKEKERKREEEKAKKEKEREEKEKRKEKEKKEKEREREKEKSKERHKKDESDSDNQDMT 905
Query: 754 AQHGYSRKSSRHK 766
HGY + + K
Sbjct: 906 DGHGYREEKKKEK 944
Score = 30.8 bits (68), Expect = 2.0
Identities = 25/92 (27%), Positives = 39/92 (42%), Gaps = 12/92 (13%)
Frame = +3
Query: 398 SDNCEMSRVHGNFSALK------------HSSNILETDSDREMGQLYGGRSSRRRQEDER 445
SDN +M+ HG K H S++ + DS+++ + R SRR D +
Sbjct: 885 SDNQDMTDGHGYREEKKKEKDKERKHRRRHQSSMDDVDSEKDEKE--ESRKSRRHGSDRK 1058
Query: 446 KQRTLDEEWNTNYKDNHKRKYRNKTSDSDSDK 477
K R + N+ DN R R+K D +
Sbjct: 1059KSR---KHANSPESDNESRHKRHKRDHGDGSR 1145
Score = 28.9 bits (63), Expect = 7.8
Identities = 23/90 (25%), Positives = 48/90 (52%)
Frame = +3
Query: 27 KERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEEREKAWIIKQQQQRDLEEEQQQ 86
KE+ +E+ R EEE+ + +E ER EE+EK K++++++ E E+++
Sbjct: 720 KEKAKEKERKR--EEEKAK-----KEKER--------EEKEKR---KEKEKKEKEREREK 845
Query: 87 LSQRDLEQHESELEEEEEEDVDDDDDGRPE 116
++ E+H+ + + + +D+ D R E
Sbjct: 846 --EKSKERHKKDESDSDNQDMTDGHGYREE 929
>AJ499968 weakly similar to GP|23495961|gb| hypothetical protein {Plasmodium
falciparum 3D7}, partial (19%)
Length = 342
Score = 47.8 bits (112), Expect = 2e-05
Identities = 31/101 (30%), Positives = 61/101 (59%), Gaps = 2/101 (1%)
Frame = -3
Query: 6 KEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEE 65
+E+++ + K K ++ RKE E+ERLE E + EE ++ E++E ER+E +E
Sbjct: 304 REEQERIAKEKEEQERKEREERERLELERIAKEKEERERKEKEEREERERLEAVVRIEKE 125
Query: 66 REKA--WIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEE 104
R++A +K+Q+++ L +E + +++ E+ +S+ EEE
Sbjct: 124 RKEAEEKALKEQEEKALLKE-GVIDKKEEEEPKSKATPEEE 5
Score = 46.6 bits (109), Expect = 4e-05
Identities = 29/91 (31%), Positives = 58/91 (62%), Gaps = 3/91 (3%)
Frame = -3
Query: 18 KERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEEREKAWIIKQQQQ 77
++ +E E+E EE+ R+ +EE++R E++E ER+E +RIA E+ E+ K++++
Sbjct: 340 EKEERERKEREEREEQERIAKEKEEQER--KEREERERLELERIAKEKEERE--RKEKEE 173
Query: 78 RDLEEEQQQLSQRDLEQHESE---LEEEEEE 105
R+ E + + + + E+ E+E L+E+EE+
Sbjct: 172 REERERLEAVVRIEKERKEAEEKALKEQEEK 80
Score = 46.2 bits (108), Expect = 5e-05
Identities = 31/106 (29%), Positives = 61/106 (57%), Gaps = 5/106 (4%)
Frame = -3
Query: 6 KEKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEE 65
KE+R+ ++ +R+E+ + EKE E + R EER+R+ LE+ E+ ER+R EE
Sbjct: 337 KEERERKEREEREEQERIAKEKEEQERKER-----EERERLELERIAKEKEERERKEKEE 173
Query: 66 RE-----KAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEED 106
RE +A + ++++++ EE+ + + E ++++EEE+
Sbjct: 172 REERERLEAVVRIEKERKEAEEKALKEQEEKALLKEGVIDKKEEEE 35
>BG644407 similar to GP|12697612|dbj contains ESTs C97999(C0353)
D15249(C0353)~similar to Arabidopsis thaliana chromosome
2 At2g01100~, partial (21%)
Length = 764
Score = 46.6 bits (109), Expect = 4e-05
Identities = 47/176 (26%), Positives = 77/176 (43%)
Frame = +2
Query: 413 LKHSSNILETDSDREMGQLYGGRSSRRRQEDERKQRTLDEEWNTNYKDNHKRKYRNKTSD 472
L+ S+N+ SD E G+ + RR+ D D + K + ++ ++
Sbjct: 302 LEASANL---KSDAEAGEKTKA-AKHRRKSDSGSDSGSDTDSEFEKKKSTRKSHKKHRKH 469
Query: 473 SDSDKEGLEEVDRKKYHEHTRKSSFNWNKDDNSRRHEEYSDVDRVTINRDNEKPHDRKGR 532
SD L+ D+K + R+SS + DD+ R ++ S+ +R R ++K H R
Sbjct: 470 HSSDLVDLDRKDKKSKRKPKRRSSDSG--DDSCREYDGDSEEERRRKKRGHKK-HRHHDR 640
Query: 533 NSQKSNRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDESESDKD 588
S ++ DS D DED G+KHH S+H R + + SES D
Sbjct: 641 YSDYNSSDSSD-------DEDV---GKKHH------SKHHKRHRRSQPNGSESSSD 760
Score = 45.4 bits (106), Expect = 8e-05
Identities = 34/139 (24%), Positives = 57/139 (40%)
Frame = +2
Query: 630 EAGSDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESDKDRGHEKYSDVDWDTMSRDNEK 689
++GSD D ++ S+++ +HR +S++ D D K
Sbjct: 392 DSGSDTDSEFEKKKSTRKSHKKHRKHHSSDLVDLDRKD---------------------K 508
Query: 690 QHDSKGKNSQKRNGDSKYQIDEAGSDEDRHRKKHLSSQMKESRHQSSDSGYGIDESDKDK 749
+ K K +GD + + S+E+R RKK K+ RH S Y +S D+
Sbjct: 509 KSKRKPKRRSSDSGDDSCREYDGDSEEERRRKKR---GHKKHRHHDRYSDYNSSDSSDDE 679
Query: 750 GEMEAQHGYSRKSSRHKRS 768
+ H S+ RH+RS
Sbjct: 680 DVGKKHH--SKHHKRHRRS 730
Score = 43.9 bits (102), Expect = 2e-04
Identities = 40/155 (25%), Positives = 60/155 (37%), Gaps = 11/155 (7%)
Frame = +2
Query: 524 EKPHDRKGRNSQKSNRDSRDWIYEAGCDEDRDGDGRKHHISRRKGSRHQSRDNSNVTDES 583
EK K R S DS D D + K +R+ +H+ +S++ D
Sbjct: 344 EKTKAAKHRRKSDSGSDS---------GSDTDSEFEKKKSTRKSHKKHRKHHSSDLVDLD 496
Query: 584 ESDK-----------DRGHEEYSDVDWDTMSRENEKQHGSNGRNSQKWNRDSRYRIYEAG 632
DK D G + + D D+ K+ G + + +R S Y ++
Sbjct: 497 RKDKKSKRKPKRRSSDSGDDSCREYDGDSEEERRRKKRGH--KKHRHHDRYSDYNSSDSS 670
Query: 633 SDEDRHRKRHHSSQRKGSRHRSRDNSNVADESESD 667
DED K+HHS K RHR R N ++ S D
Sbjct: 671 DDEDV-GKKHHSKHHK--RHR-RSQPNGSESSSDD 763
Score = 28.9 bits (63), Expect = 7.8
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 1/84 (1%)
Frame = +2
Query: 667 DKDRGHEKYSDVDWDTMSRDNEKQHDSKGKNSQ-KRNGDSKYQIDEAGSDEDRHRKKHLS 725
+K GH K+ + + ++ + K K ++ +R DS ++GSD D +K S
Sbjct: 266 EKKEGHLKWEQKLEASANLKSDAEAGEKTKAAKHRRKSDSG---SDSGSDTDSEFEKKKS 436
Query: 726 SQMKESRHQSSDSGYGIDESDKDK 749
++ +H+ S +D KDK
Sbjct: 437 TRKSHKKHRKHHSSDLVDLDRKDK 508
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 46.2 bits (108), Expect = 5e-05
Identities = 37/117 (31%), Positives = 57/117 (48%)
Frame = +3
Query: 20 RRKEIAEKERLEEEARLNDPEEERKRILLEQQEAERIERDRIAFEEREKAWIIKQQQQRD 79
+R+++ E E+E + + E+E E++E E E EE E
Sbjct: 30 KRRKVEEHNEEEDEGEVEEEEDEHD----EEEEDEHDE------EEEE------------ 143
Query: 80 LEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPEIIWLGNEIIIQKKNPNPLLL 136
EEE+++ + D E+ E E EEEEEE+ DDD++G + EI P+PLLL
Sbjct: 144 -EEEEEEEDEHDDEEEEEE-EEEEEEEEDDDEEGEED------EIDRISTLPDPLLL 290
>TC88509 similar to PIR|T01826|T01826 microfibril-associated protein homolog
T15F16.8 - Arabidopsis thaliana, partial (83%)
Length = 1306
Score = 45.4 bits (106), Expect = 8e-05
Identities = 39/114 (34%), Positives = 60/114 (52%), Gaps = 2/114 (1%)
Frame = +1
Query: 5 RKEKRKALKKLK-RKERRKEIAEKERLEEEARL-NDPEEERKRILLEQQEAERIERDRIA 62
RK+ R+ + ++ R + R+E+ E R +A + + EEE KR Q+ + E+D A
Sbjct: 265 RKDDRRLRRLIESRVDNREEVREDHRRIRQAEIVSTIEEEAKR----QEGLDLEEQDEDA 432
Query: 63 FEEREKAWIIKQQQQRDLEEEQQQLSQRDLEQHESELEEEEEEDVDDDDDGRPE 116
ER + I ++ +QRD EE Q E+ E E EEEEEE+ D + D E
Sbjct: 433 MAERRRR-IKEKLRQRDQEEALPQE-----EEEEEEEEEEEEEESDYETDSDEE 576
Score = 39.7 bits (91), Expect = 0.004
Identities = 46/198 (23%), Positives = 81/198 (40%), Gaps = 25/198 (12%)
Frame = +1
Query: 7 EKRKALKKLKRKERRKEIAEKERLEEEARLNDPEEERKRILLEQQE-----------AER 55
E+R+ +K+ R+ ++E +E EEE + EEE +E +
Sbjct: 439 ERRRRIKEKLRQRDQEEALPQEEEEEEEEEEEEEEESDYETDSDEEYTGVAMVKPVFVPK 618
Query: 56 IERDRIAFEEREKA--WIIKQQQQRDLEEEQQQLSQRDLEQHESELE-------EEEEED 106
ERD IA ER +A +++ ++R +EE + + Q +E+ + E E +D
Sbjct: 619 SERDTIAERERLEAEEEALEEARKRRMEERRNETRQIVVEEIRKDQEIQKNIELEANIDD 798
Query: 107 VDDDD---DGRPEIIWLGNEI--IIQKKNPNPLLLLHNHNHHHHHHHHQEQHEHHDNRPT 161
VD DD + W EI I + + LL + +E+ + +
Sbjct: 799 VDTDDELNEAEEYEAWKVREIGRIKRDREDREALLKEKEEVERVRNMTEEERREWERK-- 972
Query: 162 SNPFPPESPSQPLHNVAQ 179
NP P +S Q + A+
Sbjct: 973 -NPKPSQSSKQEMEIYAE 1023
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,207,084
Number of Sequences: 36976
Number of extensions: 516429
Number of successful extensions: 15858
Number of sequences better than 10.0: 679
Number of HSP's better than 10.0 without gapping: 6269
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10907
length of query: 771
length of database: 9,014,727
effective HSP length: 104
effective length of query: 667
effective length of database: 5,169,223
effective search space: 3447871741
effective search space used: 3447871741
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140549.14


