

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140547.5 - phase: 0
(603 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
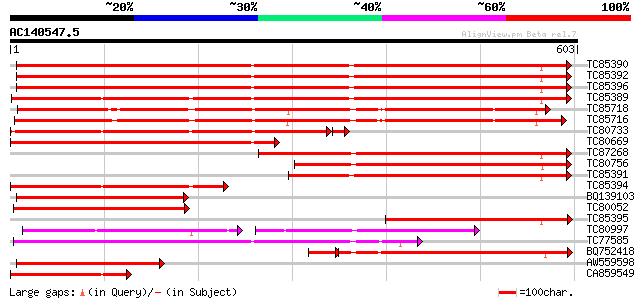
Score E
Sequences producing significant alignments: (bits) Value
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 801 0.0
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 795 0.0
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 786 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 647 0.0
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 473 e-134
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 450 e-127
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 444 e-126
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 438 e-123
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 415 e-116
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 391 e-109
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 302 3e-82
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 300 1e-81
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 285 4e-77
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 261 7e-70
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 249 2e-66
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 156 5e-66
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 239 2e-63
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 224 5e-62
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 206 2e-53
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 189 2e-48
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 801 bits (2069), Expect = 0.0
Identities = 409/604 (67%), Positives = 481/604 (78%), Gaps = 14/604 (2%)
Frame = +1
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTDTERLIGDAAKNQ+A P N
Sbjct: 160 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPTN 339
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRF D +VQ D+KLWPFKV+P DKPMIVVNYK +EK FS +EISSMVL
Sbjct: 340 TVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSAEEISSMVL 519
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K+KE+AE YLG + NAV+TVPAYFN+SQRQAT DAG I+G NV+RIINEPTAAAIAYG
Sbjct: 520 MKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIINEPTAAAIAYG 699
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 700 LDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 879
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD IS N +AL RLR+ACE+AKR LSS++QT IE+DSL GID + T+TRA
Sbjct: 880 QEFKRKNKKD--ISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYTTITRA 1053
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ K+ VH+ VLVGGST IPK+QQLL++ F
Sbjct: 1054RFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFF----NG 1221
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+
Sbjct: 1222KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPR 1401
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE VFS DNQ +LI+VYEGE +T DN LLGKFELSG PR VP I V
Sbjct: 1402NTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITV 1581
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS EE+ +M+++AE+YK+EDEE +KKV
Sbjct: 1582CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKV 1761
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
+AKN ENY Y MR + KK+E +E +I+W + NQL E DEFE K
Sbjct: 1762EAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEFEDKM 1941
Query: 594 QELE 597
+ELE
Sbjct: 1942KELE 1953
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 795 bits (2052), Expect = 0.0
Identities = 406/604 (67%), Positives = 480/604 (79%), Gaps = 14/604 (2%)
Frame = +1
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTDTERLIGDAAKNQ+A P N
Sbjct: 154 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPTN 333
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRR SD +VQ D+KLWPFKV+P DKPMIVVNYK +EK FS +EISSMVL
Sbjct: 334 TVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSAEEISSMVL 513
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K+KE+AE YLG + NAV+TVPAYFN+SQRQAT DAG I+G NVMRIINEPTAAAIAYG
Sbjct: 514 MKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAIAYG 693
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 694 LDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 873
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD IS N +AL R+R+ACE+AKR LSS++QT IE+DSL GID + T+TRA
Sbjct: 874 QEFKRKNKKD--ISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGIDFYTTITRA 1047
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ K+ V + VLVGGST IPK+QQLL++ F
Sbjct: 1048RFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQDFF----NG 1215
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+
Sbjct: 1216KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPR 1395
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE VFS DNQ +LI+VYEGE +T DN LLGKFELSG PR VP I V
Sbjct: 1396NTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITV 1575
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS E++ +M+++AE+YK+EDEE ++KV
Sbjct: 1576CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKRKV 1755
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
+AKN ENY Y MR + KK+E +E +I+W + NQL E DEFE K
Sbjct: 1756EAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEFEDKM 1935
Query: 594 QELE 597
+ELE
Sbjct: 1936KELE 1947
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 786 bits (2031), Expect = 0.0
Identities = 403/604 (66%), Positives = 480/604 (78%), Gaps = 14/604 (2%)
Frame = +2
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTD+ERLIGDAAKNQ+A P N
Sbjct: 131 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTN 310
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRR SD +VQ D+KLWPFKV+ +KPMI VNYKG+EK F+ +EISSMVL
Sbjct: 311 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLFASEEISSMVL 490
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE YLG + NAV+TVPAYFN+SQRQAT DAG IAG NV+RIINEPTAAAIAYG
Sbjct: 491 IKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 670
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +V V
Sbjct: 671 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVTHFV 850
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD IS N +AL RLR+ACE+AKR LSS++QT IE+DSL G+D + T+TRA
Sbjct: 851 QEFKRKNKKD--ISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFYTTITRA 1024
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ KS VH+ VLVGGST IPK+QQLL++ F
Sbjct: 1025RFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFF----NG 1192
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P S G+ET GGVM+VLIP+
Sbjct: 1193KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTVLIPR 1372
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE VFS DNQ +LI+VYEGE +T+DN LLGKFELSG PR VP I V
Sbjct: 1373NTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRGVPQITV 1552
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS EE+ +M+++AE+YK+EDEE +KKV
Sbjct: 1553CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKV 1732
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
+AKN ENY Y MR + K++E +E +I+W + NQLAE DEFE K
Sbjct: 1733EAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEADEFEDKM 1912
Query: 594 QELE 597
+ELE
Sbjct: 1913KELE 1924
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 647 bits (1669), Expect = 0.0
Identities = 336/611 (54%), Positives = 447/611 (72%), Gaps = 16/611 (2%)
Frame = +2
Query: 3 TRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLA 62
T+ IGIDLGT+YSC+ V++N VEII NDQGNR+TPS+V+FTD ERLIG+AAKN A
Sbjct: 146 TKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAA 325
Query: 63 KYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQE 121
P T+FD KRLIGR+F+D+ VQ+D+KL P+K+V N KP I V K G+ K FSP+E
Sbjct: 326 VNPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIV-NKDGKPYIQVRVKDGETKVFSPEE 502
Query: 122 ISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTA 181
+S+M+L+K+KE AE +LG + +AV+TVPAYFN++QRQAT DAG IAG NV RIINEPTA
Sbjct: 503 VSAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTA 682
Query: 182 AAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNN 241
AAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V +T GDTHLGG DFD
Sbjct: 683 AAIAYGLDKK---GGEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQR 853
Query: 242 LVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLH 301
++ ++L +K+ KD IS++ +ALG+LR E+AKR LSS Q +E++SL G+D
Sbjct: 854 IMEYFIKLIKKKHSKD--ISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFS 1027
Query: 302 FTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMF 361
+TRA FEE+N DLF+K M V+K + + + K+Q+ E VLVGGST IPK+QQLLK+ F
Sbjct: 1028EPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYF 1207
Query: 362 RVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVM 421
+ KEP K +NPDEAVA+GAAVQ +IL+ EG ++ +D+LLLDV P +LG+ET GGVM
Sbjct: 1208----DGKEPNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVM 1375
Query: 422 SVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRK 481
+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D LLG F+LSG PR
Sbjct: 1376TKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRG 1555
Query: 482 VPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDE 541
P I V F+VD NGIL V AEDK G +KITITN++GRLS EE+ RM+R+AE + ED+
Sbjct: 1556TPQIEVTFEVDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDK 1735
Query: 542 EVRKKVKAKNLFENYVYEMRERV---------------KKLEKVVEESIEWFERNQLAEI 586
+V++++ A+N E YVY M+ ++ +K+E V+E++EW + NQ E
Sbjct: 1736KVKERIDARNALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQTVEK 1915
Query: 587 DEFEFKKQELE 597
+EFE K +E+E
Sbjct: 1916EEFEEKLKEVE 1948
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 473 bits (1217), Expect = e-134
Identities = 273/580 (47%), Positives = 373/580 (64%), Gaps = 13/580 (2%)
Frame = +2
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDT-ERLIGDAAKNQLAKYPHN 67
IGIDLGT+ SC++V +++ N +G R TPS VAFT E L+G AK Q P N
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 442
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
T+ AKRLIGRRF D Q+++K+ P+K+V V KGQ+ +SP +I + VL
Sbjct: 443 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAW--VEAKGQQ--YSPSQIGAFVL 610
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
+K+KE AE YLG V AVITVPAYFN++QRQAT DAG+IAG V+RIINEPTAAA++YG
Sbjct: 611 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 790
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
++K E + VFDLGGGTFDVS++ I G+F+VKAT GDT LGG DFDN L++ LV
Sbjct: 791 MNK------EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 952
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLC----GGIDLHFT 303
F R DL S++ AL RLR A EKAK LSS+SQT I L + G L+ T
Sbjct: 953 NEFKRSESIDL--SKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 1126
Query: 304 VTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRV 363
+TR+ FE + +L ++ + CL + IS + E +LVGG T +PK+Q+++ +F
Sbjct: 1127LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFG- 1303
Query: 364 NDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSV 423
K PCK +NPDEAVA GAA+Q IL GD +++LLLLDV P SLG+ET GG+ +
Sbjct: 1304----KSPCKGVNPDEAVAMGAALQGGILR--GD--VKELLLLDVTPLSLGIETLGGIFTR 1459
Query: 424 LIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVP 483
LI +NT IPTKK VFS A DNQ + IKV +GE DN +LG+FEL G PR +P
Sbjct: 1460LINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMP 1639
Query: 484 NINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEV 543
I V FD+D NGI+ V+A+DK+ G +Q+ITI G LS +E++ M+++AE + +D+E
Sbjct: 1640QIEVTFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQER 1816
Query: 544 RKKVKAKNLFENYVY-------EMRERV-KKLEKVVEESI 575
+ + +N + +Y E RE++ ++ K +E+++
Sbjct: 1817KSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAV 1936
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 450 bits (1158), Expect = e-127
Identities = 269/603 (44%), Positives = 369/603 (60%), Gaps = 16/603 (2%)
Frame = +1
Query: 6 SKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDT-ERLIGDAAKNQLAKY 64
S IGIDLGT+ SC+++ ++I N +G R TPS VAF E L+G AK Q
Sbjct: 238 SDVIGIDLGTTNSCVSLMEGKNPKVIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTN 417
Query: 65 PHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISS 124
P NT+F KRLIGRRF D Q+++K+ P+K+V + +N ++ +SP +I +
Sbjct: 418 PTNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGA 585
Query: 125 MVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAI 184
VL+K+KE AE YLG ++ AV+TVPAYFN++QRQAT DAG+IAG V RIINEPTAAA+
Sbjct: 586 FVLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAAL 765
Query: 185 AYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVN 244
+YG++ K E + VFDLGGGTFDVS++ I G+F+VKAT GDT LGG DFDN L++
Sbjct: 766 SYGMNNK-----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLD 930
Query: 245 RLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSL----CGGIDL 300
LV F R DL +++ AL RLR A EKAK LSS+SQT I L + G L
Sbjct: 931 FLVSEFKRTDSIDL--AKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHL 1104
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+ T+TR+ FE + +L ++ + CL + IS V E +LVGG T +PK+Q+++ +
Sbjct: 1105NITLTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEI 1284
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F K P K +NPDEAVA GAA+Q IL GD +++LLLLDV P SLG+ET GG+
Sbjct: 1285FG-----KSPSKGVNPDEAVAMGAALQGGIL--RGD--VKELLLLDVTPLSLGIETLGGI 1437
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQD--SLLIKVYEGEHAKTEDNFLLGKFELSGCSLV 478
+ LI +NT IPTKK VFS A DNQ EG DN LG+F+L G
Sbjct: 1438FTRLISRNTTIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPA 1617
Query: 479 PRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKA 538
PR +P I V FD+D NGI+ V+A+DK+ G +Q+ITI G LS +E+ M+++AE +
Sbjct: 1618PRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSDDEINNMVKEAELHAQ 1794
Query: 539 EDEEVRKKVKAKNLFENYVY-------EMRERV-KKLEKVVEESIEWFERNQLAE-IDEF 589
D+E + + KN + +Y E RE++ ++ K +E S+ E +DE
Sbjct: 1795RDQERKALIDIKNSADTSIYSIEKSLSEYREKIPSEVAKEIENSVSDLRTAMEGESVDEI 1974
Query: 590 EFK 592
+ K
Sbjct: 1975KTK 1983
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 444 bits (1142), Expect(2) = e-126
Identities = 232/342 (67%), Positives = 278/342 (80%)
Frame = +2
Query: 1 MATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQ 60
MAT S AIGIDLGT+YSC+ + N+++EII NDQGNR TPSYVAF DTERLIGDAAKNQ
Sbjct: 173 MAT--SFAIGIDLGTTYSCVGRYANDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQ 346
Query: 61 LAKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQ 120
+A PHNTVFDAKRLIGR+FSD VQ D+K +PFKV+ + KP I V +KG+ K F+P+
Sbjct: 347 VAMNPHNTVFDAKRLIGRKFSDSEVQADMKHFPFKVI-DKGGKPNIEVEFKGENKTFTPE 523
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EIS+MVL K++E AE YLG +V NAVITVPAYFN+SQRQAT DAG IAG NV+RIINEPT
Sbjct: 524 EISAMVLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPT 703
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK EGE+NVL+FDLGGGTFDVSL+TI+EG+F+VK+T GDTHLGG DFDN
Sbjct: 704 AAAIAYGLDKK--AEGERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDN 877
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
LVN V F RK+KKDL S N +AL RLR+ACE+AKR LSSS+QT IE+DSL GID
Sbjct: 878 RLVNHFVNEFKRKHKKDL--SSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDF 1051
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFV 342
+ ++TRA FEE+ +DLF+ ++ V++ L + KI KSQVHE V
Sbjct: 1052YTSITRARFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 27.3 bits (59), Expect(2) = e-126
Identities = 11/18 (61%), Positives = 15/18 (83%)
Frame = +3
Query: 344 VGGSTTIPKIQQLLKNMF 361
VGGST IP+IQ+L+ + F
Sbjct: 1182 VGGSTRIPRIQKLISDYF 1235
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 438 bits (1127), Expect = e-123
Identities = 217/287 (75%), Positives = 251/287 (86%)
Frame = +2
Query: 1 MATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQ 60
MAT++ KAIGIDLGT+YSC+ VW+N+RVEIIPNDQGNR TPSYVAFTDTERLIGDAAKNQ
Sbjct: 98 MATKEGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQ 277
Query: 61 LAKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQ 120
+A P NTVFDAKRLIGRRFSD++VQ D+KLWPFKVVP +KPMIVVNYKG+EK F+ +
Sbjct: 278 VAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFAAE 457
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EISSMVL K++EVAE +LGH V NAV+TVPAYFN+SQRQAT DAG I+G NV+RIINEPT
Sbjct: 458 EISSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPT 637
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK R+GE+NVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN
Sbjct: 638 AAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDN 817
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQT 287
+VN V F RK KKD IS N +AL RLR+ACE+AKR LSS++QT
Sbjct: 818 RMVNHFVSEFRRKNKKD--ISGNARALRRLRTACERAKRTLSSTAQT 952
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 415 bits (1067), Expect = e-116
Identities = 214/347 (61%), Positives = 262/347 (74%), Gaps = 14/347 (4%)
Frame = +2
Query: 265 KALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRAFFEEINKDLFKKCMETV 324
+AL RLR+ACE+AKR LSS++QT IE+DSL GID + +TRA FEE+N DLF+KCME V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 325 EKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGA 384
EKCL + K+ K VH+ VLVGGST IPK+QQLL++ F KE CKSINPDEAVAYGA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNG----KELCKSINPDEAVAYGA 349
Query: 385 AVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACD 444
AVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+NT IPTKKE VFS D
Sbjct: 350 AVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSD 529
Query: 445 NQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDK 504
NQ +LI+V+EGE +T DN LLGKFELSG PR VP I VCFD+D NGIL V+AEDK
Sbjct: 530 NQPGVLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDK 709
Query: 505 TIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERV 564
T G K KITITN +GRLS E++ +M+++AE+YK+EDEE +KKV+AKN ENY Y MR +
Sbjct: 710 TTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTI 889
Query: 565 --------------KKLEKVVEESIEWFERNQLAEIDEFEFKKQELE 597
KK+E +E +I+W + NQLAE DEFE K +ELE
Sbjct: 890 KDEKIAGKLDSDDKKKIEDTIEAAIQWLDANQLAEADEFEDKMKELE 1030
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 391 bits (1005), Expect = e-109
Identities = 199/308 (64%), Positives = 240/308 (77%), Gaps = 14/308 (4%)
Frame = +3
Query: 304 VTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRV 363
+TRA FEE+N DLF+KCME VEKCL + KI KS VHE VLVGGST IPK+QQLL++ F
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 364 NDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSV 423
KE CKSINPDEAVAYGAAVQAAIL+ EGD+K++DLLLLDV P SLG+ET GGVM+
Sbjct: 183 ----KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTT 350
Query: 424 LIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVP 483
LIP+NT IPTKKE +FS DNQ +LI+V+EGE A+T+DN LLGKFEL+G PR VP
Sbjct: 351 LIPRNTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVP 530
Query: 484 NINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEV 543
INVCFD+D NGIL V+AEDKT G+K KITITN +GRLS EE+ +M++DAE+YKAEDEEV
Sbjct: 531 QINVCFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEV 710
Query: 544 RKKVKAKNLFENYVYEMRERVK--------------KLEKVVEESIEWFERNQLAEIDEF 589
+KKV+AKN ENY Y MR +K K+EK VE++I+W E NQ+AE+DEF
Sbjct: 711 KKKVEAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAEVDEF 890
Query: 590 EFKKQELE 597
E K++ELE
Sbjct: 891 EDKQKELE 914
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 302 bits (773), Expect = 3e-82
Identities = 158/316 (50%), Positives = 218/316 (68%), Gaps = 15/316 (4%)
Frame = +1
Query: 297 GIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQL 356
G+ +TRA FEE+N DLF+K M V+K + + + K+Q+ E VLVGGST IPK+QQL
Sbjct: 4 GVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQL 183
Query: 357 LKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVET 416
LK+ F + KEP K +NPDEAVA+GAAVQ +IL+ EG + +D+LLLDV P +LG+ET
Sbjct: 184 LKDYF----DGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIET 351
Query: 417 KGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCS 476
GGVM+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D LLGKF+LSG
Sbjct: 352 VGGVMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIP 531
Query: 477 LVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERY 536
PR P I V F+VD NGIL V AEDK G +KITITN++GRLS EE+ RM+R+AE +
Sbjct: 532 PAPRGTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEF 711
Query: 537 KAEDEEVRKKVKAKNLFENYVYEMRERV---------------KKLEKVVEESIEWFERN 581
ED++V++++ A+N E YVY M+ +V +K+E V+E++EW + N
Sbjct: 712 AEEDKKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDN 891
Query: 582 QLAEIDEFEFKKQELE 597
Q E +E+E K +E+E
Sbjct: 892 QSVEKEEYEEKLKEVE 939
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 300 bits (768), Expect = 1e-81
Identities = 153/232 (65%), Positives = 185/232 (78%), Gaps = 1/232 (0%)
Frame = +1
Query: 2 ATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQL 61
AT+ IGIDLGT+YSC+ V++N VEII NDQGNR+TPS+V+FTD ERLIG+AAKN
Sbjct: 277 ATKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLA 456
Query: 62 AKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQ 120
A P T+FD KRLIGR+F D+ VQ+D+KL P+K+V N KP I V K G+ K FSP+
Sbjct: 457 AVNPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NRDGKPYIQVRVKDGETKVFSPE 633
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EIS+M+L K+KE AE +LG + +AV+TVPAYFN++QRQAT DAG IAG NV RIINEPT
Sbjct: 634 EISAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 813
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTH 232
AAAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V AT GDTH
Sbjct: 814 AAAIAYGLDKK---GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 285 bits (729), Expect = 4e-77
Identities = 137/183 (74%), Positives = 160/183 (86%)
Frame = +2
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTD+ERLIGDAAKNQ+A P N
Sbjct: 80 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPIN 259
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRFSD +VQ D+KLWPFK++ +KP+I VNYKG++K F+ +EISSMVL
Sbjct: 260 TVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEFAAEEISSMVL 439
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE YLG + NAV+TVPAYFN+SQRQAT DAG IAG NVMRIINEPTAAAIAYG
Sbjct: 440 MKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAIAYG 619
Query: 188 LDK 190
LDK
Sbjct: 620 LDK 628
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 261 bits (666), Expect = 7e-70
Identities = 128/187 (68%), Positives = 152/187 (80%)
Frame = +3
Query: 5 KSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKY 64
K AIGIDLGT+YSC+AV +N ++II ND G R TPS+VAF D+ER+IGDAA N A
Sbjct: 96 KKVAIGIDLGTTYSCVAVCKNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAAFNIAASN 275
Query: 65 PHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISS 124
P NT+FDAKRLIGR+FSD VQ D+KLWPFKV+ + DKPMIVVNY +EKHF+ +EISS
Sbjct: 276 PTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAAEEISS 455
Query: 125 MVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAI 184
MVL K++E+AET+LG V + VITVPAYFN+SQRQ+T DAG IAG NVMRIINEPTAAAI
Sbjct: 456 MVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEPTAAAI 635
Query: 185 AYGLDKK 191
AYG + K
Sbjct: 636 AYGFNTK 656
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 249 bits (636), Expect = 2e-66
Identities = 127/213 (59%), Positives = 154/213 (71%), Gaps = 14/213 (6%)
Frame = +3
Query: 400 EDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHA 459
+DLLLLDV P SLG+ET GGVM+VLIP+NT IPTKKE VFS DNQ +LI+VYEGE A
Sbjct: 3 QDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERA 182
Query: 460 KTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEG 519
+T DN LLGKFELSG PR VP I VCFD+D NGIL V+AEDKT G K KITITN +G
Sbjct: 183 RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKG 362
Query: 520 RLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERV--------------K 565
RLS EE+ +M+ +AE+YKAEDEE +KKV AKN ENY Y MR + K
Sbjct: 363 RLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKK 542
Query: 566 KLEKVVEESIEWFERNQLAEIDEFEFKKQELEN 598
K++ ++ +I+W + NQLAE DEF+ K +ELE+
Sbjct: 543 KIDDAIDAAIQWLDSNQLAEADEFQDKMKELES 641
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 156 bits (394), Expect(2) = 5e-66
Identities = 84/239 (35%), Positives = 135/239 (56%), Gaps = 1/239 (0%)
Frame = +1
Query: 262 ENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRAFFEEINKDLFKKCM 321
E K++ LR A + A LS+ S +++D L + + V RA FEE+NK++F+KC
Sbjct: 763 EEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCE 939
Query: 322 ETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVA 381
+ +CL + K+ +++ +LVGG + IP+++ L+ N+ ++ E K INP E
Sbjct: 940 SLIIQCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLCKIT----EVYKGINPLEGAV 1107
Query: 382 YGAAVQAAILNTEGDKKIE-DLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFS 440
GA ++ A+ + D DLL + P ++G+ G +IP+NT +P +K+ +F+
Sbjct: 1108CGATMEGAVASGISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFT 1287
Query: 441 IACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEV 499
DNQ LI VYEGE K E+N LLG F++ G P+ VP I+VC D+D +L V
Sbjct: 1288TIHDNQTEALILVYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 114 bits (284), Expect(2) = 5e-66
Identities = 77/243 (31%), Positives = 126/243 (51%), Gaps = 9/243 (3%)
Frame = +3
Query: 14 GTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAK--NQLAKYPHNTVFD 71
GTS +AVW + VE++ N + ++ S+V F D G ++ ++ +T+F+
Sbjct: 3 GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFN 182
Query: 72 AKRLIGRRFSDQTVQQDIKLWPFKVVP-NHKDKPMIVVNYKGQEKHFSPQEISSMVLSKL 130
KRLIGR +D V L PF V + +P I + +P+E+ +M L +L
Sbjct: 183 MKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLVEL 359
Query: 131 KEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGLDK 190
+ + ET+L + N V+TVP F+ Q A +AG +V+R++ EPTA A+ YG +
Sbjct: 360 RLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQQQ 539
Query: 191 K------MWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVN 244
+ M EK L+F++G G DV++ G+ Q+KA G T +GG D N++
Sbjct: 540 QKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNMMR 716
Query: 245 RLV 247
L+
Sbjct: 717 HLL 725
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 239 bits (611), Expect = 2e-63
Identities = 142/448 (31%), Positives = 232/448 (51%), Gaps = 13/448 (2%)
Frame = +1
Query: 5 KSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKY 64
K +G D G +AV R ++++ ND+ R TP+ V F D +R IG A
Sbjct: 235 KMSVVGFDFGNESCIVAVARQRGIDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMN 414
Query: 65 PHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISS 124
P N++ KRLIG++F+D +Q+D+K PF V P+I Y G+ + F+ ++
Sbjct: 415 PKNSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFG 594
Query: 125 MVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAI 184
M+LS LKE+A+ L V + I +P YF + QR++ DA IAG + + +I+E TA A+
Sbjct: 595 MMLSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATAL 774
Query: 185 AYGLDKKMWREGE-KNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLV 243
AYG+ K E E NV D+G + V + +G V + D LGG DFD L
Sbjct: 775 AYGIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALF 954
Query: 244 NRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFT 303
+ F +YK D + +N +A RLR+ACEK K++LS++ + + ++ L D+
Sbjct: 955 HHFAAKFKEEYKID--VYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGF 1128
Query: 304 VTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRV 363
+ R FE+++ + ++ +EK L E ++ +H +VG + +P I ++L F+
Sbjct: 1129IKRDDFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK- 1305
Query: 364 NDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGV--------- 414
KEP +++N E VA GAA+Q AIL+ K+ + + + PFS+ +
Sbjct: 1306----KEPRRTMNASECVARGAALQCAILSP--TFKVREFQVNESFPFSVSLSWKYSGSDA 1467
Query: 415 ---ETKGGVMSVLIPKNTMIPTKKENVF 439
E+ +++ PK IP+ K F
Sbjct: 1468PDSESDNKQSTIVFPKGNPIPSSKVLTF 1551
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 224 bits (572), Expect(2) = 5e-62
Identities = 120/265 (45%), Positives = 176/265 (66%), Gaps = 16/265 (6%)
Frame = -1
Query: 350 IPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMP 409
IPK+Q L++ F K+ K INPDEAVA+GAAVQA +L+ G++ E+++L+DV P
Sbjct: 900 IPKVQSLIEEYFGG----KKASKGINPDEAVAFGAAVQAGVLS--GEEGTEEIVLMDVNP 739
Query: 410 FSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGK 469
+LG+ET GGVM+ LI +NT IPT+K +FS A DNQ +LI+V+EGE + T+DN LGK
Sbjct: 738 LTLGIETTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGK 559
Query: 470 FELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRM 529
FEL+G PR VP I V F++D NGIL+V+A DK G ++ ITITN +GRL+ EE+ RM
Sbjct: 558 FELTGIPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRM 379
Query: 530 MRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERVKKLE---------------KVVEES 574
+ +AE+Y ED+ R++++A+N ENY + ++ +V E + V+E+
Sbjct: 378 VEEAEKYAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETILEAVKET 199
Query: 575 IEWFERN-QLAEIDEFEFKKQELEN 598
+W E N A ++FE +K++L N
Sbjct: 198 NDWLEENGATANTEDFEEQKEKLSN 124
Score = 32.0 bits (71), Expect(2) = 5e-62
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = -3
Query: 318 KKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPK 352
++ VE+ L + K+ K V + VLVGGST P+
Sbjct: 997 RRLSSPVEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 206 bits (525), Expect = 2e-53
Identities = 103/159 (64%), Positives = 127/159 (79%), Gaps = 2/159 (1%)
Frame = +1
Query: 8 AIGIDLGTSYSCIAVW--RNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYP 65
A+GIDLGT+YSC+AVW NRVEII ND+G+++TPS+VAFTD +RL+G AAK+Q A P
Sbjct: 55 AVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAAKDQAAINP 234
Query: 66 HNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSM 125
NTVFDAKRLIGR+FSD VQ+DI LWPFKVV DKPMI + +KGQEK +EISS+
Sbjct: 235 QNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLLCAEEISSI 414
Query: 126 VLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDA 164
VL+ ++E+AE YL V NA ITVPAYFN++QR+AT DA
Sbjct: 415 VLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 189 bits (481), Expect = 2e-48
Identities = 92/129 (71%), Positives = 109/129 (84%)
Frame = +3
Query: 1 MATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQ 60
M ++ KA+GIDLGT+YSC+ VW+N+RVEII NDQGNR TPSYVAFTDTERLIGDAAKNQ
Sbjct: 144 MDPQEGKAVGIDLGTTYSCVGVWQNDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQ 323
Query: 61 LAKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQ 120
+A P+NTVFDAKRLIGRRF+DQ VQ D+K WPFKV+ + KP I V YKG+ K F+P+
Sbjct: 324 VAMNPYNTVFDAKRLIGRRFADQEVQSDMKHWPFKVI-DKAAKPYIQVEYKGETKQFTPE 500
Query: 121 EISSMVLSK 129
EISSMVL+K
Sbjct: 501 EISSMVLTK 527
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,960,136
Number of Sequences: 36976
Number of extensions: 199966
Number of successful extensions: 1125
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 1054
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1061
length of query: 603
length of database: 9,014,727
effective HSP length: 102
effective length of query: 501
effective length of database: 5,243,175
effective search space: 2626830675
effective search space used: 2626830675
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140547.5


