

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140032.3 - phase: 0 /pseudo
(617 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
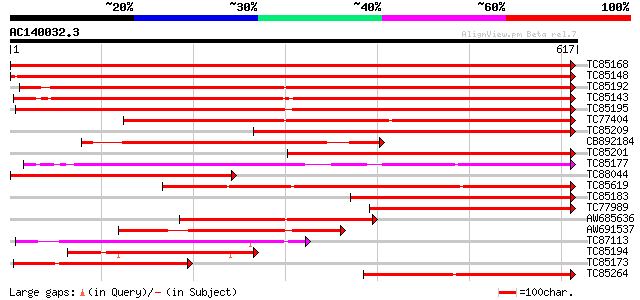
Score E
Sequences producing significant alignments: (bits) Value
TC85168 similar to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12... 1241 0.0
TC85148 homologue to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.... 1146 0.0
TC85192 homologue to PIR|T12142|T12142 lipoxygenase (EC 1.13.11.... 839 0.0
TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12... 815 0.0
TC85195 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 806 0.0
TC77404 homologue to GP|493730|emb|CAA55318.1| lipoxygenase {Pis... 736 0.0
TC85209 homologue to PIR|T06454|T06454 probable lipoxygenase (EC... 626 e-180
CB892184 homologue to PIR|T06454|T06 probable lipoxygenase (EC 1... 461 e-130
TC85201 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11... 446 e-126
TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 446 e-125
TC88044 similar to PIR|T06454|T06454 probable lipoxygenase (EC 1... 395 e-110
TC85619 similar to PIR|T11578|T11578 probable lipoxygenase (EC 1... 388 e-108
TC85183 homologue to SP|P09918|LOX3_PEA Seed lipoxygenase-3 (EC ... 360 e-100
TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Coryl... 311 4e-85
AW685636 similar to SP|P38414|LOX1 Lipoxygenase (EC 1.13.11.12).... 297 7e-81
AW691537 similar to PIR|S56655|S56 lipoxygenase (EC 1.13.11.12) ... 291 5e-79
TC87113 weakly similar to GP|8649004|emb|CAB94852.1 lipoxygenase... 263 2e-70
TC85194 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 248 5e-66
TC85173 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11... 246 2e-65
TC85264 similar to GP|14589309|emb|CAC43237. lipoxygenase {Sesba... 236 3e-62
>TC85168 similar to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12) - garden
pea, partial (98%)
Length = 2890
Score = 1241 bits (3210), Expect = 0.0
Identities = 615/615 (100%), Positives = 615/615 (100%)
Frame = +2
Query: 1 MFGILNNKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTAT 60
MFGILNNKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTAT
Sbjct: 62 MFGILNNKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTAT 241
Query: 61 AFLGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGI 120
AFLGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGI
Sbjct: 242 AFLGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGI 421
Query: 121 PGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQ 180
PGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQ
Sbjct: 422 PGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQ 601
Query: 181 TPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYP 240
TPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYP
Sbjct: 602 TPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYP 781
Query: 241 YPRRVRSGRKPTRKDPKSEKPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVI 300
YPRRVRSGRKPTRKDPKSEKPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVI
Sbjct: 782 YPRRVRSGRKPTRKDPKSEKPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVI 961
Query: 301 FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVI 360
FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVI
Sbjct: 962 FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVI 1141
Query: 361 KVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLG 420
KVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLG
Sbjct: 1142KVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLG 1321
Query: 421 DITVEEALNGKKLFLLDYHDAFMPYLERININAKAYATRTILFLKDDGTLKPIAIELSLP 480
DITVEEALNGKKLFLLDYHDAFMPYLERININAKAYATRTILFLKDDGTLKPIAIELSLP
Sbjct: 1322DITVEEALNGKKLFLLDYHDAFMPYLERININAKAYATRTILFLKDDGTLKPIAIELSLP 1501
Query: 481 HSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIA 540
HSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIA
Sbjct: 1502HSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIA 1681
Query: 541 TNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRD 600
TNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRD
Sbjct: 1682TNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRD 1861
Query: 601 WVFTDQALPADLIKR 615
WVFTDQALPADLIKR
Sbjct: 1862WVFTDQALPADLIKR 1906
>TC85148 homologue to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12) -
garden pea, complete
Length = 2962
Score = 1146 bits (2964), Expect = 0.0
Identities = 559/615 (90%), Positives = 592/615 (95%)
Frame = +2
Query: 1 MFGILNNKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTAT 60
MFGI + KGQKIKGTVVLMPKNVLDFNAITS+GKGGV++ AGN+IGGVT IVGGVVDTAT
Sbjct: 65 MFGIFD-KGQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTAT 241
Query: 61 AFLGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGI 120
AFLGRNVSMQLISATKTDA+GKGLVGKETFLSKHLPQLPTLGARQDAFS+FFEYDANFGI
Sbjct: 242 AFLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGI 421
Query: 121 PGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQ 180
PGAFYIRNYTQAEFFLV VTLEDIPNR SVQF CNSWVYNFKSYK +RIFFTND YLPSQ
Sbjct: 422 PGAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQ 601
Query: 181 TPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYP 240
TPAPLNH+REEELQ LRGDGTGERKE DR+YDYD+YNDLGNPDGGDALVRP+LGGSST+P
Sbjct: 602 TPAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHP 781
Query: 241 YPRRVRSGRKPTRKDPKSEKPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVI 300
YPRRVR+GRKPTRKD KSEKPG IYVPRDENFGHLKSSDFLMYGIKSLSQ+V+PL +SVI
Sbjct: 782 YPRRVRTGRKPTRKDLKSEKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVI 961
Query: 301 FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVI 360
FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEI RTDGEQ LKFPPP VI
Sbjct: 962 FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVI 1141
Query: 361 KVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLG 420
+VSKSAW TDEEFGREM+AGVNPNVIRLLQEFPPKSTLD TVYGDQNSTITKEHL TNLG
Sbjct: 1142RVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLG 1321
Query: 421 DITVEEALNGKKLFLLDYHDAFMPYLERININAKAYATRTILFLKDDGTLKPIAIELSLP 480
DITVEEAL+GK+LFLLDYHDAFMPYLERIN+NAKAYATRTILFL+DDGTLKP+AIELSLP
Sbjct: 1322DITVEEALDGKRLFLLDYHDAFMPYLERINLNAKAYATRTILFLQDDGTLKPLAIELSLP 1501
Query: 481 HSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIA 540
HSNGVQYG+ESKVFLPA EGVESTIW+LAKAHVIVNDSCYHQL+SHWLNTHAV+EPFIIA
Sbjct: 1502HSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSHWLNTHAVMEPFIIA 1681
Query: 541 TNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRD 600
TNRHLSVLHPINKLLFPH+RDTININGLARQ+LINAGGIIEQTF PGPNS+EISS VY++
Sbjct: 1682TNRHLSVLHPINKLLFPHYRDTININGLARQALINAGGIIEQTFCPGPNSIEISSAVYKN 1861
Query: 601 WVFTDQALPADLIKR 615
WVFTDQALPADLIKR
Sbjct: 1862WVFTDQALPADLIKR 1906
>TC85192 homologue to PIR|T12142|T12142 lipoxygenase (EC 1.13.11.12) 1 -
fava bean, complete
Length = 2918
Score = 839 bits (2168), Expect = 0.0
Identities = 415/607 (68%), Positives = 494/607 (81%), Gaps = 2/607 (0%)
Frame = +1
Query: 11 KIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLGRNVSMQ 70
KIKGT+VLM KNVLD N++T K II G G + DT T+FLGR++ +Q
Sbjct: 49 KIKGTLVLMQKNVLDINSLTDPTK---------IIDGALDGFGSIFDTLTSFLGRSICLQ 201
Query: 71 LISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPGAFYIRNYT 130
LIS+TK G+G +GKE +L + + LPTLG +Q AFS+ FEYD+NFGIPGAF I+N+
Sbjct: 202 LISSTKIGLTGEGKLGKEAYLKEAINNLPTLGDKQTAFSIEFEYDSNFGIPGAFKIKNFM 381
Query: 131 QAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAPLNHYRE 190
EF LVS+TL+DIPN ++ F+CNSWVYN K+Y DRIFF N+T+LPS+TPAPL +YR+
Sbjct: 382 STEFLLVSLTLDDIPNVGTIHFVCNSWVYNAKNYLTDRIFFANNTFLPSETPAPLVYYRQ 561
Query: 191 EELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRRVRSGRK 250
EL+TLRGDGTGERKE DRVYDYD+YNDLG+PD G + RPVLGGSS+ PYPRR R+GRK
Sbjct: 562 LELKTLRGDGTGERKEWDRVYDYDVYNDLGDPDKGQSYARPVLGGSSSLPYPRRGRTGRK 741
Query: 251 PTRKDPKSE-KPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDLNFTPNE 309
PT DP SE + +Y+PRDE FGHLKSSDFL+YG+KS+SQ+VIPL +SV FD NFTPNE
Sbjct: 742 PTATDPNSESRSSSVYIPRDEAFGHLKSSDFLVYGLKSVSQDVIPLIQSV-FDTNFTPNE 918
Query: 310 FDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKSAWMT 369
FDSFD+V L+EGGIKLPTDILSQISPLP L EIFRTDGEQ LKFP P VI+VSKSAWMT
Sbjct: 919 FDSFDDVLDLYEGGIKLPTDILSQISPLPVLSEIFRTDGEQFLKFPTPKVIQVSKSAWMT 1098
Query: 370 DEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEEALN 429
D+EFGRE++AGVNP +IR LQEFPPKS LD+ VYGD STIT+EH+ NL +TV+EA+
Sbjct: 1099DDEFGREILAGVNPGLIRSLQEFPPKSKLDSAVYGDHTSTITREHIQLNLDGLTVDEAIQ 1278
Query: 430 GKKLFLLDYHDAFMPYLERIN-INAKAYATRTILFLKDDGTLKPIAIELSLPHSNGVQYG 488
KKLFLL++HD +PYL IN + KAYA+RT+LFLK DGTLKP+AIELSLPH G Q+G
Sbjct: 1279NKKLFLLEHHDTIIPYLRLINSTSTKAYASRTVLFLKSDGTLKPLAIELSLPHPQGDQFG 1458
Query: 489 SESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVL 548
S V+LPA EGVE+T+WLLAKA+VIVNDSC+HQL+SHWLNTHAVVEPF+IATNR LSVL
Sbjct: 1459VVSNVYLPAIEGVEATVWLLAKAYVIVNDSCFHQLVSHWLNTHAVVEPFVIATNRQLSVL 1638
Query: 549 HPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFTDQAL 608
HPI KLL PH+RDT+NIN LAR SL+NA GIIE+TFL G ++EISS VY+DWVFTDQAL
Sbjct: 1639HPIYKLLHPHYRDTMNINALARSSLVNADGIIEKTFLWGGYAMEISSKVYKDWVFTDQAL 1818
Query: 609 PADLIKR 615
PADLIKR
Sbjct: 1819PADLIKR 1839
>TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12) - kidney
bean, partial (93%)
Length = 3009
Score = 815 bits (2106), Expect = 0.0
Identities = 413/615 (67%), Positives = 486/615 (78%), Gaps = 4/615 (0%)
Frame = +1
Query: 5 LNNKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLG 64
L K QKIKGTVVLM KN LD N I K G AG G V +VGGV+D ATA LG
Sbjct: 52 LFKKKQKIKGTVVLMCKNALDLNDI----KAGPSLGAG--FGLVRDVVGGVIDGATAILG 213
Query: 65 RNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPGAF 124
R+V +QLISATKTD G G VG + FL K +P LPTL ARQDAFSV+FE+D +FGIPGAF
Sbjct: 214 RSVGLQLISATKTDRMGNGFVGDQFFLEKRIPFLPTLAARQDAFSVYFEWDNDFGIPGAF 393
Query: 125 YIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAP 184
YI+NYTQ EF+LVS+TLEDIP +S+ F+CNSWVYN K Y+KDRIFF N YLP + PAP
Sbjct: 394 YIKNYTQGEFYLVSLTLEDIPGHDSITFLCNSWVYNAKQYRKDRIFFANKPYLPHEMPAP 573
Query: 185 LNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRR 244
L +YR EL TLRGDGT +R+E DR+YDYD+YNDLGNPD L RP+LGGSS +PYPRR
Sbjct: 574 LVNYRHVELDTLRGDGTAKREEWDRIYDYDVYNDLGNPDKSRDLARPILGGSSKFPYPRR 753
Query: 245 VRSGRKPTRKDPKSEKPG--VIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFD 302
R+ RKPT+ D SE+P YVPRDE FGHLK +DFL +G+K L+QNVIP F++ FD
Sbjct: 754 GRTCRKPTKTDSASERPATDTNYVPRDEVFGHLKQADFLGFGLKGLNQNVIPKFRN-FFD 930
Query: 303 LNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKV 362
+ EFD+F+EVR LFEGG+KLPTDI+S ISP+ +KEIFRTDGE LK+PPPHVI+
Sbjct: 931 FD---KEFDNFEEVRCLFEGGVKLPTDIISAISPIAMVKEIFRTDGENFLKYPPPHVIQA 1101
Query: 363 SKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDI 422
KSAWMTDEEF REM+AGVNP VI+ LQ FPP+S LD VYGDQ STITKEHL N +
Sbjct: 1102DKSAWMTDEEFAREMLAGVNPCVIQRLQVFPPESKLDPNVYGDQTSTITKEHLEINTDGL 1281
Query: 423 TVEEALNGKKLFLLDYHDAFMPYLERININ-AKAYATRTILFLKDDGTLKPIAIELSLPH 481
TVE+A+ ++LF+LDYHDAFM YL+ IN K+YATRTILFLKDDGTLKP+AIELSLPH
Sbjct: 1282TVEKAIQDERLFILDYHDAFMSYLKYINKPIPKSYATRTILFLKDDGTLKPLAIELSLPH 1461
Query: 482 SNGVQYGSESKVFLPAD-EGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIA 540
+G +YG+ SKV LP + GV+ TIW LAKA+V+VND+C+HQL+SHWLNTH V+EPFIIA
Sbjct: 1462PDGEKYGAVSKVLLPPEGHGVQRTIWQLAKAYVVVNDACFHQLMSHWLNTHCVIEPFIIA 1641
Query: 541 TNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRD 600
TNR LSV+HPI+KLL PH+RDT+NIN LAR SLI+ GGIIEQ FLPGP +VE+SS VY+D
Sbjct: 1642TNRCLSVVHPIHKLLQPHYRDTMNINALARSSLISGGGIIEQAFLPGPYAVEMSSAVYKD 1821
Query: 601 WVFTDQALPADLIKR 615
WVF DQALPADLIKR
Sbjct: 1822WVFPDQALPADLIKR 1866
>TC85195 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (98%)
Length = 2903
Score = 806 bits (2081), Expect = 0.0
Identities = 389/611 (63%), Positives = 480/611 (77%), Gaps = 2/611 (0%)
Frame = +3
Query: 7 NKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLGRN 66
N+G+K+KGTVVLM KNVLD N +TS+ V G IGG G VG V+DTATAFLGRN
Sbjct: 87 NRGEKLKGTVVLMQKNVLDVNELTSIKSNPVGGIIGGAIGGAFGAVGTVLDTATAFLGRN 266
Query: 67 VSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPGAFYI 126
V+++LISAT D +GKG VGK+TFL + +PTLG +Q+A+SV FE+ ++ GIPGAFYI
Sbjct: 267 VALKLISATSADGSGKGKVGKQTFLEGVITSIPTLGDKQNAYSVHFEWGSDMGIPGAFYI 446
Query: 127 RNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAPLN 186
N+ Q EFFLVS+TLED+PN ++ F+CNSWVYN K YK DRIFF N TYLPS+TPAPL
Sbjct: 447 ENFLQHEFFLVSLTLEDVPNHGTINFVCNSWVYNDKKYKSDRIFFANKTYLPSETPAPLV 626
Query: 187 HYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRRVR 246
+YR+EEL+TLRGDG GERKE +R+YDYD+YNDLG PD L RPVLGGSST PYPRR R
Sbjct: 627 YYRQEELKTLRGDGKGERKEWERIYDYDVYNDLGEPDSKPTLGRPVLGGSSTLPYPRRGR 806
Query: 247 SGRKPTRKDPKSE-KPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDLNF 305
+GRKP +KDPKSE + G +Y+PRDE FGH KSSDFL + +KS SQN+IP KSV+
Sbjct: 807 TGRKPAKKDPKSESRSGTVYLPRDEAFGHTKSSDFLAFILKSASQNIIPSLKSVV----- 971
Query: 306 TPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKS 365
EF++F++VR L++GGIKLPT+ LS +SP+P E+FR+DG LKF PP V++V S
Sbjct: 972 -SKEFNNFEDVRSLYDGGIKLPTNFLSNVSPIPLFTELFRSDGASTLKFSPPKVVQVDHS 1148
Query: 366 AWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVE 425
AWMTDEEF REM+AGVNP++I+ L EFP KS LD+ YGD STITKE L NLG ITVE
Sbjct: 1149AWMTDEEFAREMIAGVNPHIIKKLTEFPHKSKLDSQKYGDNTSTITKEQLEPNLGGITVE 1328
Query: 426 EALNGKKLFLLDYHDAFMPYLERIN-INAKAYATRTILFLKDDGTLKPIAIELSLPHSNG 484
+A+ KL++LD++D PYL +IN KAYA RTILFL++DGTLKP+AIELS PH +
Sbjct: 1329QAIQNNKLYILDHYDIVYPYLRKINATETKAYAARTILFLQNDGTLKPLAIELSKPHPDD 1508
Query: 485 VQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRH 544
YG S+V+ PA EGVE++IWLLAKA+V+VNDSC+HQL+SHWLNTHAVVEPFIIATNRH
Sbjct: 1509DSYGPVSEVYFPASEGVEASIWLLAKAYVVVNDSCHHQLVSHWLNTHAVVEPFIIATNRH 1688
Query: 545 LSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFT 604
LS +HP++KLL PH+RDT+NIN LAR L+NA GIIE TFL G ++E+S++ YRDWVFT
Sbjct: 1689LSTVHPVHKLLLPHYRDTMNINSLARNVLVNAEGIIESTFLWGGYALEMSAVAYRDWVFT 1868
Query: 605 DQALPADLIKR 615
+Q LP DL+KR
Sbjct: 1869EQGLPNDLLKR 1901
>TC77404 homologue to GP|493730|emb|CAA55318.1| lipoxygenase {Pisum
sativum}, partial (85%)
Length = 2463
Score = 736 bits (1899), Expect = 0.0
Identities = 360/493 (73%), Positives = 416/493 (84%), Gaps = 2/493 (0%)
Frame = +3
Query: 125 YIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAP 184
YI+NY Q EF+L S+TLED+PN +++F+CNSWVYN K YK RIFF N +YLPS+TP+P
Sbjct: 3 YIKNYMQVEFYLKSLTLEDVPNHGTIRFVCNSWVYNAKLYKSPRIFFANKSYLPSETPSP 182
Query: 185 LNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRR 244
L YREEELQ LRGDGTGERK +R+YDYD+YNDLGNPD G++ RPVLGGSST+PYPRR
Sbjct: 183 LVKYREEELQNLRGDGTGERKLHERIYDYDVYNDLGNPDHGESFARPVLGGSSTHPYPRR 362
Query: 245 VRSGRKPTRKDPKSEKPGV-IYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDL 303
R+GR PTRKDP SEKP IYVPRDENFGHLKSSDFL YGIKS+SQNV+P F+S FDL
Sbjct: 363 GRTGRYPTRKDPNSEKPATEIYVPRDENFGHLKSSDFLTYGIKSVSQNVLPAFESA-FDL 539
Query: 304 NFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVS 363
NFTP EFDSF +VR LFEGGIKLP D++S ISPLP +KE+FRTDGE VLKFP PHV+KVS
Sbjct: 540 NFTPREFDSFQDVRDLFEGGIKLPLDVISTISPLPVIKELFRTDGENVLKFPTPHVVKVS 719
Query: 364 KSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDIT 423
KSAWMTDEEF REM+AGVNP +IR L+EFPPKS LD TVYGDQ S IT + A L T
Sbjct: 720 KSAWMTDEEFAREMLAGVNPCMIRGLKEFPPKSNLDPTVYGDQTSKITAD--ALELDGST 893
Query: 424 VEEALNGKKLFLLDYHDAFMPYLERIN-INAKAYATRTILFLKDDGTLKPIAIELSLPHS 482
V+EAL G +LF+LDYHD F+P+L RIN +AKAYATRTILFLK+DG LKP+AIELSLPH
Sbjct: 894 VDEALAGGRLFILDYHDTFIPFLRRINETSAKAYATRTILFLKEDGNLKPVAIELSLPHP 1073
Query: 483 NGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATN 542
+G + G S+V LPA+EG E TIWLLAKA+V+VNDSCYHQL+SHWLNTHAVVEPF+IATN
Sbjct: 1074DGDKSGVVSQVILPANEGGERTIWLLAKAYVVVNDSCYHQLMSHWLNTHAVVEPFVIATN 1253
Query: 543 RHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWV 602
R LSV+HPI KLL PH+RDT+NIN LAR+SLINA GIIE+TFLP +VE+SS VY++WV
Sbjct: 1254RQLSVIHPIYKLLSPHYRDTMNINALARESLINANGIIERTFLPSKYAVEMSSAVYKNWV 1433
Query: 603 FTDQALPADLIKR 615
F DQALPADLIKR
Sbjct: 1434FPDQALPADLIKR 1472
>TC85209 homologue to PIR|T06454|T06454 probable lipoxygenase (EC
1.13.11.12) - garden pea, partial (68%)
Length = 2002
Score = 626 bits (1615), Expect = e-180
Identities = 305/350 (87%), Positives = 329/350 (93%)
Frame = +3
Query: 266 VPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDLNFTPNEFDSFDEVRGLFEGGIK 325
VPRDENFGHLKSSDFL YGIKSLSQ+V+PLFKSVIFDLNFTPNEFDSFDEVR LFEGGI+
Sbjct: 3 VPRDENFGHLKSSDFLTYGIKSLSQDVLPLFKSVIFDLNFTPNEFDSFDEVRDLFEGGIE 182
Query: 326 LPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKSAWMTDEEFGREMVAGVNPNV 385
LPT ILS+ISPLP LKEIFRTDGEQVLKFPPPHVI+VSKSAWMTDEEFGREMVAGVNP V
Sbjct: 183 LPTHILSKISPLPVLKEIFRTDGEQVLKFPPPHVIRVSKSAWMTDEEFGREMVAGVNPCV 362
Query: 386 IRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEEALNGKKLFLLDYHDAFMPY 445
IRLLQEFPPKSTLD VYGDQ S + KEHL NLG +TVE+ALNG++LF+LDYHDAFMP+
Sbjct: 363 IRLLQEFPPKSTLDIAVYGDQTSILKKEHLEINLGGLTVEKALNGQRLFILDYHDAFMPF 542
Query: 446 LERININAKAYATRTILFLKDDGTLKPIAIELSLPHSNGVQYGSESKVFLPADEGVESTI 505
LE+IN NAKAYATRTILFLKDDGTLKP+AIELSLPH NGV+YG+ESKV LPAD+GV+STI
Sbjct: 543 LEKINKNAKAYATRTILFLKDDGTLKPVAIELSLPHPNGVKYGAESKVILPADQGVDSTI 722
Query: 506 WLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLHPINKLLFPHFRDTINI 565
WLLAKAHVIVNDSCYHQL+SHWLNTHAVVEPFIIATNRHLSVLHPINKLL PHFRDTINI
Sbjct: 723 WLLAKAHVIVNDSCYHQLMSHWLNTHAVVEPFIIATNRHLSVLHPINKLLDPHFRDTINI 902
Query: 566 NGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFTDQALPADLIKR 615
NGLAR +LINA GIIE+TFLPGPNSVE+SS Y++WVFTDQALPADLIKR
Sbjct: 903 NGLARNALINADGIIEETFLPGPNSVEMSSAAYKNWVFTDQALPADLIKR 1052
>CB892184 homologue to PIR|T06454|T06 probable lipoxygenase (EC 1.13.11.12) -
garden pea, partial (31%)
Length = 839
Score = 461 bits (1186), Expect = e-130
Identities = 237/331 (71%), Positives = 256/331 (76%), Gaps = 1/331 (0%)
Frame = +1
Query: 79 ANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPGAFYIRNYTQA-EFFLV 137
ANGKGLVGKE F GAFYIRNY QA EFFLV
Sbjct: 1 ANGKGLVGKEVFF------------------------------GAFYIRNYMQAHEFFLV 90
Query: 138 SVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAPLNHYREEELQTLR 197
SVT +DIPN ESV+F+CNSW+YNFK+YKKDRIFFTNDTYLPSQTPAPL +YR+EELQ LR
Sbjct: 91 SVTFDDIPNHESVEFVCNSWIYNFKNYKKDRIFFTNDTYLPSQTPAPLVYYRQEELQNLR 270
Query: 198 GDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRRVRSGRKPTRKDPK 257
GDGTG+RKE +RVYDYD+YNDLG+PD L RPVLGGSST+PYPRRVRSGRKPT+KDPK
Sbjct: 271 GDGTGQRKEWERVYDYDVYNDLGDPDEDVKLARPVLGGSSTHPYPRRVRSGRKPTKKDPK 450
Query: 258 SEKPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDLNFTPNEFDSFDEVR 317
SE+PGV+YVPRDENFGHLKSSDFL YGIKSLSQ+V+PLFKSVIFDLNFTPNEFDSFDEVR
Sbjct: 451 SERPGVMYVPRDENFGHLKSSDFLTYGIKSLSQDVLPLFKSVIFDLNFTPNEFDSFDEVR 630
Query: 318 GLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKSAWMTDEEFGREM 377
LFEGGI+LPT ILS+ISPLP LKEIFR TDEEFGREM
Sbjct: 631 DLFEGGIELPTHILSKISPLPVLKEIFR-----------------------TDEEFGREM 741
Query: 378 VAGVNPNVIRLLQEFPPKSTLDTTVYGDQNS 408
VAGVNP VIRLLQEFPPKSTLD VYGDQ S
Sbjct: 742 VAGVNPCVIRLLQEFPPKSTLDIAVYGDQTS 834
>TC85201 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11.12).
[Lentil] {Lens culinaris}, partial (49%)
Length = 1275
Score = 446 bits (1148), Expect = e-126
Identities = 212/314 (67%), Positives = 259/314 (81%), Gaps = 1/314 (0%)
Frame = +1
Query: 303 LNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKV 362
L F EF++F++VR L+EGGI+LPTDILS++SP+P KE+FRTDGE LKFPPP VI+V
Sbjct: 7 LQFHEPEFNTFEDVRSLYEGGIRLPTDILSELSPIPLFKELFRTDGEAALKFPPPKVIQV 186
Query: 363 SKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDI 422
SAWMTDEEF REM+AGVNP++I+ L EFPPKS LDT ++G+ STITKEHL N+ +
Sbjct: 187 DHSAWMTDEEFAREMIAGVNPHIIKKLLEFPPKSKLDTQLFGNNTSTITKEHLQPNMVGV 366
Query: 423 TVEEALNGKKLFLLDYHDAFMPYLERININ-AKAYATRTILFLKDDGTLKPIAIELSLPH 481
TVE+A+ KLF+LD+HD PYL +IN KAYATRTILFL++DGTLKP+AIELS PH
Sbjct: 367 TVEQAIQNNKLFILDHHDPLFPYLRKINATETKAYATRTILFLQNDGTLKPLAIELSRPH 546
Query: 482 SNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIAT 541
G G S V+LPA EGVE++IWLLAKA+V+VNDSCYHQL+SHWLNTHAVVEPFIIAT
Sbjct: 547 PQGDSLGPVSNVYLPASEGVEASIWLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFIIAT 726
Query: 542 NRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDW 601
NRHLSV+HPI+KLL PH+RDT+NIN LAR L+NAGG++E TFL G +VE+S++VY+DW
Sbjct: 727 NRHLSVVHPIHKLLLPHYRDTMNINSLARTILVNAGGVMELTFLWGDYAVEMSAVVYKDW 906
Query: 602 VFTDQALPADLIKR 615
FT+Q LP DLIKR
Sbjct: 907 NFTEQGLPNDLIKR 948
>TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (75%)
Length = 2646
Score = 446 bits (1147), Expect = e-125
Identities = 265/604 (43%), Positives = 354/604 (57%), Gaps = 4/604 (0%)
Frame = +1
Query: 16 VVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLGRNVSMQLISAT 75
VVLM KN L++ I G V+ G+ + ++DT+ F +LISA+
Sbjct: 61 VVLMKKNALNYKVI---GGNTVVENEGDTVRPT------LLDTSVGF-------KLISAS 192
Query: 76 KTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDAN-FGIPGAFYIRNYTQAEF 134
K DA GKG VGKETF+ + +P LG Q+AFS+ FE+D N GIPGAFY++N+TQ E
Sbjct: 193 KADATGKGKVGKETFMDGFVTSIPNLGDIQNAFSIHFEWDPNHMGIPGAFYVKNFTQDEI 372
Query: 135 FLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAPLNHYREEELQ 194
FLVS+TLED+ + E+ FICNSW+YN + Y+ +RIFF N YL +TPAPL +YR+EEL
Sbjct: 373 FLVSLTLEDVESHETTNFICNSWIYNAEKYQTERIFFANKAYLLRETPAPLLYYRQEELN 552
Query: 195 TLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRRVRSGRKPTRK 254
TLRGDG + N A + LG +R
Sbjct: 553 TLRGDGLESVRNGTEYMIMMSTMIWVNQTKILACIVQFLGDQLPCLTLEGEELAENTSRN 732
Query: 255 DPKSEKPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDLNFTPNEFDSFD 314
+ I+ ++ N L++ L+ + N+ L + L EF+SF
Sbjct: 733 IQRLRVGVTIFTFQEMN*LSLRNRQTLLLTPSNQYLNMRHLN*RSLVRLQNDQVEFNSFH 912
Query: 315 EVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKSAWMTDEEFG 374
+V LF G + KF P VI+ ++AWMTDEEF
Sbjct: 913 DVLSLFAG---------------------------EHPKFSTPLVIQEDRTAWMTDEEFA 1011
Query: 375 REMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGD-ITVEEALNGKKL 433
REM+AGVNPNVI+ + +TI K+H + D ++VE+ + +L
Sbjct: 1012REMIAGVNPNVIKKV---------------GNTTTINKKHFEPYMQDGVSVEQTIKDLRL 1146
Query: 434 FLLDYHDAFMPYLERININ--AKAYATRTILFLKDDGTLKPIAIELSLPHSNGVQYGSES 491
+++DY DA +PYL +IN N AKAYA+ T+L L+DDGTLKPI+IEL +PH +G G +
Sbjct: 1147YVVDYQDAILPYLRKINANGAAKAYASTTLLSLQDDGTLKPISIELHVPHPDGD--GIVT 1320
Query: 492 KVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLHPI 551
+ PA EGV+++IW LAKA+ +VND+CYHQLISHWL+THA VEPFIIATNRHLSV+HPI
Sbjct: 1321TSYTPATEGVDASIWRLAKAYAVVNDACYHQLISHWLHTHASVEPFIIATNRHLSVVHPI 1500
Query: 552 NKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFTDQALPAD 611
+KLL PH+R+T+NIN AR LI AGGIIE T+L G S+E+SS VY+DWVF DQALP D
Sbjct: 1501HKLLLPHYRNTMNINANARDVLIKAGGIIESTYLFGSYSMELSSDVYKDWVFPDQALPND 1680
Query: 612 LIKR 615
LIKR
Sbjct: 1681LIKR 1692
>TC88044 similar to PIR|T06454|T06454 probable lipoxygenase (EC 1.13.11.12)
- garden pea, partial (28%)
Length = 804
Score = 395 bits (1015), Expect = e-110
Identities = 190/248 (76%), Positives = 219/248 (87%), Gaps = 2/248 (0%)
Frame = +2
Query: 1 MFGILN-NKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTA 59
MFGI + + QKIKGTVVLMPKNVLDFNA+TSV +GG ++ AGN++ +VGG++D A
Sbjct: 59 MFGIFDRDHSQKIKGTVVLMPKNVLDFNAVTSVKQGGFLDAAGNVLDAAGSLVGGIIDGA 238
Query: 60 TAFLGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFG 119
TAFLGRNVSM+LISATKTDANGKGLVGKE FL KH+P LPTLGARQDAFS+ F++DA+FG
Sbjct: 239 TAFLGRNVSMRLISATKTDANGKGLVGKEVFLEKHVPTLPTLGARQDAFSIHFDWDADFG 418
Query: 120 IPGAFYIRNYTQA-EFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLP 178
IPGAFYIRNY QA EFFLVSVT +DIPN ESV+F+CNSW+YNFK+YKKDRIFFTNDTYLP
Sbjct: 419 IPGAFYIRNYMQAHEFFLVSVTFDDIPNHESVEFVCNSWIYNFKNYKKDRIFFTNDTYLP 598
Query: 179 SQTPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSST 238
SQTPAPL +YR+EELQ LRGDGTG+RKE +RVYDYD+YNDLG+PD L RPVLGGSST
Sbjct: 599 SQTPAPLVYYRQEELQNLRGDGTGQRKEWERVYDYDVYNDLGDPDEDVKLARPVLGGSST 778
Query: 239 YPYPRRVR 246
+PYPRRVR
Sbjct: 779 HPYPRRVR 802
>TC85619 similar to PIR|T11578|T11578 probable lipoxygenase (EC 1.13.11.12)
CPRD46 drought-inducible - cowpea, partial (75%)
Length = 2452
Score = 388 bits (997), Expect = e-108
Identities = 216/456 (47%), Positives = 276/456 (60%), Gaps = 7/456 (1%)
Frame = +3
Query: 167 DRIFFTNDTYLPSQTPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGD 226
++ F +YLPS+TP L REEEL+TLRG+G GERK DR+YDYD+YNDLG+PD
Sbjct: 96 NQFFSFEQSYLPSETPEGLIRLREEELKTLRGNGQGERKSFDRIYDYDVYNDLGDPDSDP 275
Query: 227 ALVRPVLGGSSTYPYPRRVRSGRKPTRKDPKSEK-PGVIYVPRDENFGHLKSSDFLMYGI 285
L RPVLGG +PYPRR R+GR DP SEK +YVPRDE+F LK F +
Sbjct: 276 ELKRPVLGGKE-HPYPRRCRTGRPRCDTDPLSEKRSNNVYVPRDESFSELKQLSFSANTL 452
Query: 286 KSLSQNVIPLFKSVIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQI--SPLPALKEI 343
+S ++P+ K+ + D N F F + LF G LP S LP L +
Sbjct: 453 QSAFHALLPVVKTAVIDKNLG---FPLFSAIDDLFNEGFNLPPQAEKGFLASVLPRLVRL 623
Query: 344 FRTDGEQVLKFPPPHVIKVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVY 403
+L+F P + + W DEEFGR+ +AG+NP I+L+ E+P KS LD +Y
Sbjct: 624 VNEARNDILRFETPATMDKDRFFWFRDEEFGRQTIAGLNPCCIQLVTEWPLKSKLDPKIY 803
Query: 404 GDQNSTITKEHLATNL-GDITVEEALNGKKLFLLDYHDAFMPYLERINI--NAKAYATRT 460
G S IT E + + G TVEEA+ KKLF+LDY+D F+P +E + Y +RT
Sbjct: 804 GPAESAITTEMIEQQIRGYATVEEAIKQKKLFILDYYDFFLPLVEEVRKLEGTTLYGSRT 983
Query: 461 ILFLKDDGTLKPIAIELSLPHSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCY 520
+ FL +D TL+P+AIEL+ P G E VF PA +W LAKAHV+ +DS Y
Sbjct: 984 LFFLNEDSTLRPVAIELARPPIGGKPEWRE--VFTPAWHSTGVWLWRLAKAHVLAHDSGY 1157
Query: 521 HQLISHWLNTHAVVEPFIIATNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGII 580
HQL+SHWL TH EP+IIATNR LS +HPI KLL PHFR T+ IN LAR++LINA GII
Sbjct: 1158HQLVSHWLRTHCATEPYIIATNRQLSAMHPIYKLLLPHFRYTMEINALAREALINADGII 1337
Query: 581 EQTFLPGPNSVEISSIVY-RDWVFTDQALPADLIKR 615
E +F P S+ +SSI Y + W F QALP DLI R
Sbjct: 1338ESSFTPKQLSLLVSSIAYDKHWQFDLQALPNDLIHR 1445
>TC85183 homologue to SP|P09918|LOX3_PEA Seed lipoxygenase-3 (EC
1.13.11.12). [Garden pea] {Pisum sativum}, partial (57%)
Length = 1623
Score = 360 bits (924), Expect = e-100
Identities = 179/246 (72%), Positives = 205/246 (82%), Gaps = 1/246 (0%)
Frame = +3
Query: 371 EEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEEALNG 430
EEF REM+AGVNPNVI LQEFPP+S LD+ VYGD S ITKEHL NL +TVEEA+
Sbjct: 3 EEFAREMLAGVNPNVICCLQEFPPRSKLDSQVYGDHTSKITKEHLEPNLEGLTVEEAIQN 182
Query: 431 KKLFLLDYHDAFMPYLERININ-AKAYATRTILFLKDDGTLKPIAIELSLPHSNGVQYGS 489
KKLFLLD+HD+ MPYL RIN KAYATRTILFL D TLKP+AIELSLPH +G ++G+
Sbjct: 183 KKLFLLDHHDSIMPYLRRINSTPTKAYATRTILFLSSDKTLKPLAIELSLPHPDGDEHGA 362
Query: 490 ESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLH 549
S V+ PA EGVESTIWLLAKA+V+VNDSCYHQL+SHWLNTHAVVEPF+IATNRHLS LH
Sbjct: 363 VSHVYQPALEGVESTIWLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFVIATNRHLSYLH 542
Query: 550 PINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFTDQALP 609
PI KLL+PH+RDT+NIN LARQSL+N GGIIE+TFL G S+E+SS VY++W QALP
Sbjct: 543 PIYKLLYPHYRDTMNINSLARQSLVNDGGIIEKTFLWGRYSMEMSSKVYKNWTLPGQALP 722
Query: 610 ADLIKR 615
ADLIKR
Sbjct: 723 ADLIKR 740
>TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Corylus
avellana}, partial (54%)
Length = 1763
Score = 311 bits (798), Expect = 4e-85
Identities = 146/225 (64%), Positives = 187/225 (82%), Gaps = 1/225 (0%)
Frame = +3
Query: 392 FPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEEALNGKKLFLLDYHDAFMPYLERIN- 450
FPP S LD +VYGDQNS+I +H+ +L +T++EAL KL++LD+HDA MPYL RIN
Sbjct: 12 FPPASKLDPSVYGDQNSSIQAKHIENSLDGLTIDEALESDKLYILDHHDALMPYLSRINS 191
Query: 451 INAKAYATRTILFLKDDGTLKPIAIELSLPHSNGVQYGSESKVFLPADEGVESTIWLLAK 510
N K YATRT+LFL+DDGTLKP+AIELSLPH G Q+G+ SKVF P+ EGV +T+W LAK
Sbjct: 192 TNTKTYATRTLLFLQDDGTLKPLAIELSLPHPQGEQHGAVSKVFTPSHEGVAATVWQLAK 371
Query: 511 AHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLHPINKLLFPHFRDTININGLAR 570
A+ VNDS YHQL+SHWL THAV+EPFIIATNR LS+LHPI+KLL PHF+DT++IN LAR
Sbjct: 372 AYAAVNDSGYHQLVSHWLFTHAVIEPFIIATNRQLSLLHPIHKLLKPHFKDTMHINALAR 551
Query: 571 QSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFTDQALPADLIKR 615
+LINAGG++E+T PG ++E+S++VY++WVFT+QALPA+L+KR
Sbjct: 552 HTLINAGGVLEKTVFPGKFALEMSAVVYKNWVFTEQALPANLLKR 686
>AW685636 similar to SP|P38414|LOX1 Lipoxygenase (EC 1.13.11.12). [Lentil]
{Lens culinaris}, partial (25%)
Length = 665
Score = 297 bits (761), Expect = 7e-81
Identities = 142/217 (65%), Positives = 175/217 (80%), Gaps = 1/217 (0%)
Frame = +3
Query: 185 LNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRR 244
L +YR+EEL+TLRGDGTGERKE +R+YDYD+YNDLG PD L R +LGGSS +PYPRR
Sbjct: 3 LVYYRQEELKTLRGDGTGERKEWERIYDYDVYNDLGEPDSKPQLARQILGGSSDFPYPRR 182
Query: 245 VRSGRKPTRKDPKSE-KPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDL 303
R+GR+ T+ DPKSE + +Y+PRDE+FGH KSSDFL+Y +KS SQNVIP +SV+ L
Sbjct: 183 GRTGRRKTKTDPKSETRSPTVYLPRDESFGHTKSSDFLVYILKSASQNVIPQLRSVV-TL 359
Query: 304 NFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVS 363
EF++F +VR L+EGGIKLPTD+LSQISP+P KE+FR+DGEQ LKFPPP VI+V
Sbjct: 360 QLNNPEFNTFQDVRSLYEGGIKLPTDVLSQISPIPLFKELFRSDGEQALKFPPPKVIQVD 539
Query: 364 KSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDT 400
SAW TDEEF REM+AGVNP++I+ L EFPPKS LD+
Sbjct: 540 HSAWQTDEEFAREMIAGVNPHIIKKLSEFPPKSKLDS 650
>AW691537 similar to PIR|S56655|S56 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (25%)
Length = 668
Score = 291 bits (745), Expect = 5e-79
Identities = 144/248 (58%), Positives = 175/248 (70%), Gaps = 1/248 (0%)
Frame = +2
Query: 119 GIPGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLP 178
GIPGAFYI N+ Q EFFLVS+TLED+PN ++ F+CNSWVYN K YK DRIFF N
Sbjct: 2 GIPGAFYIENFLQHEFFLVSLTLEDVPNHGTINFVCNSWVYNDKKYKSDRIFFAN----- 166
Query: 179 SQTPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSST 238
+TLRGDG GERKE +R+YDYD+YNDLG PD L RPVLGGSST
Sbjct: 167 ---------------KTLRGDGKGERKEWERIYDYDVYNDLGEPDSKPTLGRPVLGGSST 301
Query: 239 YPYPRRVRSGRKPTRKDPKSE-KPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFK 297
PYPRR R+GRKP +KDPKSE + G +Y+PRDE FGH KSSDFL + +KS SQN+IP K
Sbjct: 302 LPYPRRGRTGRKPAKKDPKSESRSGTVYLPRDEAFGHTKSSDFLAFILKSASQNIIPSLK 481
Query: 298 SVIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPP 357
SV+ EF++F++VR L++GGIKLPT+ LS +SP+P E+FR+DG LKF PP
Sbjct: 482 SVV------SKEFNNFEDVRSLYDGGIKLPTNFLSNVSPIPLFTELFRSDGASTLKFSPP 643
Query: 358 HVIKVSKS 365
V +V S
Sbjct: 644 KVGQVDHS 667
>TC87113 weakly similar to GP|8649004|emb|CAB94852.1 lipoxygenase {Prunus
dulcis}, partial (32%)
Length = 1032
Score = 263 bits (671), Expect = 2e-70
Identities = 152/330 (46%), Positives = 193/330 (58%), Gaps = 9/330 (2%)
Frame = +1
Query: 7 NKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLGRN 66
N +KG VVLM K++LDF+ I S V+D FLG+
Sbjct: 103 NYKNSVKGKVVLMKKSLLDFHDIKS----------------------NVLDRIHEFLGKG 216
Query: 67 VSMQLISATKTD-ANG-KGLVGKETFLSKHLPQLPTLGARQDA-FSVFFEYD-ANFGIPG 122
VS+QLISAT D A G KG GK L + + + +L D FSV F++D G+PG
Sbjct: 217 VSLQLISATAPDPAKGLKGKHGKVACLERWMSSISSLTTATDTEFSVTFDWDHEKMGVPG 396
Query: 123 AFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTP 182
AF IRN ++F+L +VTL+DIP + F+CNSWVY Y DR+FF N YLP TP
Sbjct: 397 AFLIRNNHHSQFYLKTVTLDDIPGHGPITFVCNSWVYPTHRYTHDRVFFANKAYLPCDTP 576
Query: 183 APLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYP 242
L REEEL TLRG G G+ E DRVYDY YNDLG PD G RPV+GGS +PYP
Sbjct: 577 EALRKLREEELGTLRGKGIGKLNEWDRVYDYACYNDLGTPDNGPDYARPVIGGSQKFPYP 756
Query: 243 RRVRSGRKPTRKDPKSEK-----PGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFK 297
RR R+ R T+ DPK+E IYVPRDE FGH+K SDFL Y +KS++Q ++P +
Sbjct: 757 RRGRTSRPHTKTDPKTESRLHLLNLNIYVPRDEQFGHVKFSDFLAYSLKSVTQVLLPELR 936
Query: 298 SVIFDLNFTPNEFDSFDEVRGLFEGGIKLP 327
S+ + T NEFD+F +V ++EG LP
Sbjct: 937 SL---CDKTINEFDTFQDVLDIYEGSFNLP 1017
>TC85194 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (14%)
Length = 887
Score = 248 bits (633), Expect = 5e-66
Identities = 125/217 (57%), Positives = 159/217 (72%), Gaps = 10/217 (4%)
Frame = +3
Query: 64 GRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDAN---FGI 120
GR+V+++LIS + D NGKG VG+ T+L + T Q F++ FE+D
Sbjct: 141 GRSVALRLISDSTVDGNGKGKVGERTYLGSLIASDET----QFHFNITFEWDTEKLGAPS 308
Query: 121 PGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQ 180
PGAF+I N+ Q+E FLVS+TL D+PN ++ F CNSW+YN K+Y +RIFFTN TYL S+
Sbjct: 309 PGAFFIENFNQSELFLVSLTLNDVPNHGTINFNCNSWIYNAKNYHSERIFFTNKTYLLSE 488
Query: 181 TPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSST-- 238
TPAPL +YR+EEL+TLRGDGTGERKE DR+YDYD+YNDLG PD L RPVLGGSST
Sbjct: 489 TPAPLVYYRQEELKTLRGDGTGERKEWDRIYDYDVYNDLGEPDKEAGLGRPVLGGSSTLE 668
Query: 239 ----YPYPRRVRSGRKPTRKDPKSE-KPGVIYVPRDE 270
+PYPRRVRSGR+P++KDPK+E + G IY+PRDE
Sbjct: 669 GSDGFPYPRRVRSGREPSKKDPKTESRTGRIYIPRDE 779
>TC85173 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11.12).
[Lentil] {Lens culinaris}, partial (22%)
Length = 643
Score = 246 bits (628), Expect = 2e-65
Identities = 121/196 (61%), Positives = 151/196 (76%), Gaps = 1/196 (0%)
Frame = +1
Query: 5 LNNKGQKIKGTVVLMPKNVLDFNAITSV-GKGGVINTAGNIIGGVTGIVGGVVDTATAFL 63
L ++GQK+KGTVVLM KN LD NA+T+ G+I A I+G V G ++DTAT+FL
Sbjct: 64 LFDRGQKLKGTVVLMQKNTLDINALTAAKSPTGIIGGAFGIVGDVAG---NILDTATSFL 234
Query: 64 GRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPGA 123
GR+++++LISAT DA GKG VGKETFL L +PTLG +Q+AFS+ FE+D+N GIPGA
Sbjct: 235 GRSIALRLISATSADAAGKGKVGKETFLEGLLTSIPTLGDKQNAFSIHFEWDSNMGIPGA 414
Query: 124 FYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPA 183
FYI N+ Q EFFLVS+TLED+PN S+ F+CNSW+YN K YK DRIFF N TYLPS+TPA
Sbjct: 415 FYIDNFMQGEFFLVSLTLEDVPNHGSIHFVCNSWIYNSKKYKTDRIFFANKTYLPSETPA 594
Query: 184 PLNHYREEELQTLRGD 199
PL +YR+ EL TLRGD
Sbjct: 595 PLVYYRQ*ELNTLRGD 642
>TC85264 similar to GP|14589309|emb|CAC43237. lipoxygenase {Sesbania
rostrata}, partial (51%)
Length = 1774
Score = 236 bits (601), Expect = 3e-62
Identities = 122/233 (52%), Positives = 157/233 (67%), Gaps = 3/233 (1%)
Frame = +2
Query: 386 IRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEEALNGKKLFLLDYHDAFMPY 445
+R L FPP S LD +YG Q S + KEH+ L +TV+EA++ KLF++DYHD ++P+
Sbjct: 23 LRRLTVFPPVSKLDPEIYGPQESALKKEHILNQLNGMTVQEAIDQNKLFIIDYHDIYLPF 202
Query: 446 LERINI--NAKAYATRTILFLKDDGTLKPIAIELSLPHSNGVQYGSESKVFLPADEGVES 503
LERIN K+YATRTI +L GTLKP+AIELSLP S +V PA + +
Sbjct: 203 LERINALDGRKSYATRTIYYLTPLGTLKPVAIELSLPPSG--PNTRSKRVVTPALDATTN 376
Query: 504 TIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLHPINKLLFPHFRDTI 563
+W+LAKAHV ND+ HQL HWL THA +EPFI++ +R LS +HPI KLL PH R T+
Sbjct: 377 WMWMLAKAHVCSNDAGVHQLAHHWLRTHACMEPFILSAHRQLSAMHPIFKLLDPHMRYTL 556
Query: 564 NINGLARQSLINAGGIIEQTFLPGPNSVEISSIVYR-DWVFTDQALPADLIKR 615
IN LARQSLINA G+IE F PG ++EISS Y+ +W F +LP DLI+R
Sbjct: 557 EINALARQSLINADGVIESCFTPGRYAMEISSAAYKTNWRFDQDSLPQDLIRR 715
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,844,011
Number of Sequences: 36976
Number of extensions: 258982
Number of successful extensions: 1407
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 1319
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1338
length of query: 617
length of database: 9,014,727
effective HSP length: 102
effective length of query: 515
effective length of database: 5,243,175
effective search space: 2700235125
effective search space used: 2700235125
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140032.3


