

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140032.2 - phase: 0 /pseudo
(611 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
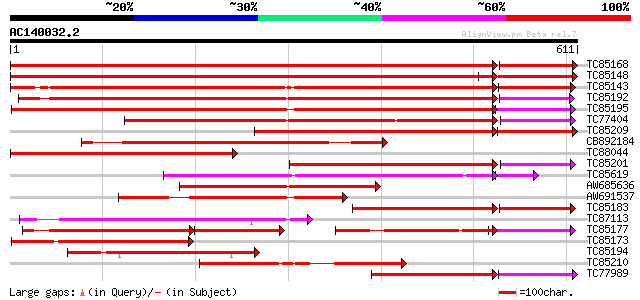
Score E
Sequences producing significant alignments: (bits) Value
TC85168 similar to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12... 978 0.0
TC85148 homologue to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.... 1061 0.0
TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12... 665 0.0
TC85192 homologue to PIR|T12142|T12142 lipoxygenase (EC 1.13.11.... 691 0.0
TC85195 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 661 0.0
TC77404 homologue to GP|493730|emb|CAA55318.1| lipoxygenase {Pis... 584 e-177
TC85209 homologue to PIR|T06454|T06454 probable lipoxygenase (EC... 453 e-149
CB892184 homologue to PIR|T06454|T06 probable lipoxygenase (EC 1... 443 e-125
TC88044 similar to PIR|T06454|T06454 probable lipoxygenase (EC 1... 398 e-111
TC85201 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11... 305 9e-92
TC85619 similar to PIR|T11578|T11578 probable lipoxygenase (EC 1... 286 2e-81
AW685636 similar to SP|P38414|LOX1 Lipoxygenase (EC 1.13.11.12).... 295 5e-80
AW691537 similar to PIR|S56655|S56 lipoxygenase (EC 1.13.11.12) ... 285 4e-77
TC85183 homologue to SP|P09918|LOX3_PEA Seed lipoxygenase-3 (EC ... 227 5e-73
TC87113 weakly similar to GP|8649004|emb|CAB94852.1 lipoxygenase... 263 1e-70
TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 168 7e-69
TC85173 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11... 245 4e-65
TC85194 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 243 1e-64
TC85210 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 211 9e-55
TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Coryl... 190 3e-52
>TC85168 similar to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12) - garden
pea, partial (98%)
Length = 2890
Score = 978 bits (2527), Expect(2) = 0.0
Identities = 478/526 (90%), Positives = 505/526 (95%), Gaps = 1/526 (0%)
Frame = +2
Query: 1 MFGIFD-KGQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTAT 59
MFGI + KGQKIKGTVVLMPKNVLDFNAITS+GKGGV++ AGN+IGGVT IVGGVVDTAT
Sbjct: 62 MFGILNNKGQKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTAT 241
Query: 60 AFLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGI 119
AFLGRNVSMQLISATKTDA+GKGLVGKETFLSKHLPQLPTLGARQDAFS+FFEYDANFGI
Sbjct: 242 AFLGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGI 421
Query: 120 PGAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQ 179
PGAFYIRNYTQAEFFLV VTLEDIPNR SVQF CNSWVYNFKSYK +RIFFTND YLPSQ
Sbjct: 422 PGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQ 601
Query: 180 TPAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHP 239
TPAPLNH+REEELQ LRGDGTGERKE DR+YDYD+YNDLGNPDGGDALVRP+LGGSST+P
Sbjct: 602 TPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYP 781
Query: 240 YPRRVRTGRKPTRKDLKSEKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVI 299
YPRRVR+GRKPTRKD KSEKPG IYVPRDENFGHLKSSDFLMYGIKSLSQ+V+PL +SVI
Sbjct: 782 YPRRVRSGRKPTRKDPKSEKPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVI 961
Query: 300 FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVI 359
FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEI RTDGEQ LKFPPP VI
Sbjct: 962 FDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVI 1141
Query: 360 RVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLG 419
+VSKSAW TDEEFGREM+AGVNPNVIRLLQEFPPKSTLD TVYGDQNSTITKEHL TNLG
Sbjct: 1142KVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLG 1321
Query: 420 DITVEEALDGKRLFLLDYHDAFMPYLERINLNAKAYATRTILFLQDDGTLKPLAIELSLP 479
DITVEEAL+GK+LFLLDYHDAFMPYLERIN+NAKAYATRTILFL+DDGTLKP+AIELSLP
Sbjct: 1322DITVEEALNGKKLFLLDYHDAFMPYLERININAKAYATRTILFLKDDGTLKPIAIELSLP 1501
Query: 480 HSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
HSNGVQYG+ESKVFLPA EGVESTIW+LAKAHVIVNDSCYHQL+SH
Sbjct: 1502HSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISH 1639
Score = 129 bits (324), Expect(2) = 0.0
Identities = 64/83 (77%), Positives = 67/83 (80%)
Frame = +1
Query: 529 TIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH*DFF 588
TIHHSNK* SKC+SP+** FI P*HNKYQWTCST SNQCW HYRANI +RTKF DFF
Sbjct: 1666 TIHHSNK*ASKCSSPN**TFISSFP*HNKYQWTCSTISNQCWWHYRANIFARTKFSRDFF 1845
Query: 589 CSLQELGFH*SSITS*SYQERIS 611
SLQ LGFH SIT *SYQER S
Sbjct: 1846 NSLQGLGFHRPSITG*SYQERTS 1914
>TC85148 homologue to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12) -
garden pea, complete
Length = 2962
Score = 1061 bits (2744), Expect = 0.0
Identities = 525/525 (100%), Positives = 525/525 (100%)
Frame = +2
Query: 1 MFGIFDKGQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATA 60
MFGIFDKGQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATA
Sbjct: 65 MFGIFDKGQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATA 244
Query: 61 FLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIP 120
FLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIP
Sbjct: 245 FLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIP 424
Query: 121 GAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQT 180
GAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQT
Sbjct: 425 GAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQT 604
Query: 181 PAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPY 240
PAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPY
Sbjct: 605 PAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPY 784
Query: 241 PRRVRTGRKPTRKDLKSEKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIF 300
PRRVRTGRKPTRKDLKSEKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIF
Sbjct: 785 PRRVRTGRKPTRKDLKSEKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIF 964
Query: 301 DLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIR 360
DLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIR
Sbjct: 965 DLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIR 1144
Query: 361 VSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGD 420
VSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGD
Sbjct: 1145VSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGD 1324
Query: 421 ITVEEALDGKRLFLLDYHDAFMPYLERINLNAKAYATRTILFLQDDGTLKPLAIELSLPH 480
ITVEEALDGKRLFLLDYHDAFMPYLERINLNAKAYATRTILFLQDDGTLKPLAIELSLPH
Sbjct: 1325ITVEEALDGKRLFLLDYHDAFMPYLERINLNAKAYATRTILFLQDDGTLKPLAIELSLPH 1504
Query: 481 SNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
SNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH
Sbjct: 1505SNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 1639
Score = 177 bits (448), Expect = 1e-44
Identities = 91/107 (85%), Positives = 93/107 (86%), Gaps = 1/107 (0%)
Frame = +1
Query: 506 MLAKAHV-IVNDSCYHQLMSHCDGTIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCST 564
ML+ H +V SC DGTIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCST
Sbjct: 1615 MLSSTHEPLVEYSC-------SDGTIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCST 1773
Query: 565 SSNQCWRHYRANILSRTKFH*DFFCSLQELGFH*SSITS*SYQERIS 611
SSNQCWRHYRANILSRTKFH*DFFCSLQELGFH*SSITS*SYQERIS
Sbjct: 1774 SSNQCWRHYRANILSRTKFH*DFFCSLQELGFH*SSITS*SYQERIS 1914
>TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12) - kidney
bean, partial (93%)
Length = 3009
Score = 665 bits (1715), Expect(2) = 0.0
Identities = 343/529 (64%), Positives = 401/529 (74%), Gaps = 4/529 (0%)
Frame = +1
Query: 1 MFGIFDKGQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATA 60
MF +F K QKIKGTVVLM KN LD N I K G AG G V +VGGV+D ATA
Sbjct: 43 MFPLFKKKQKIKGTVVLMCKNALDLNDI----KAGPSLGAG--FGLVRDVVGGVIDGATA 204
Query: 61 FLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIP 120
LGR+V +QLISATKTD G G VG + FL K +P LPTL ARQDAFS++FE+D +FGIP
Sbjct: 205 ILGRSVGLQLISATKTDRMGNGFVGDQFFLEKRIPFLPTLAARQDAFSVYFEWDNDFGIP 384
Query: 121 GAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQT 180
GAFYI+NYTQ EF+LV +TLEDIP S+ F CNSWVYN K Y+ +RIFF N YLP +
Sbjct: 385 GAFYIKNYTQGEFYLVSLTLEDIPGHDSITFLCNSWVYNAKQYRKDRIFFANKPYLPHEM 564
Query: 181 PAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPY 240
PAPL ++R EL LRGDGT +R+EWDRIYDYDVYNDLGNPD L RPILGGSS PY
Sbjct: 565 PAPLVNYRHVELDTLRGDGTAKREEWDRIYDYDVYNDLGNPDKSRDLARPILGGSSKFPY 744
Query: 241 PRRVRTGRKPTRKDLKSEKPG--AIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSV 298
PRR RT RKPT+ D SE+P YVPRDE FGHLK +DFL +G+K L+Q+V+P ++
Sbjct: 745 PRRGRTCRKPTKTDSASERPATDTNYVPRDEVFGHLKQADFLGFGLKGLNQNVIPKFRN- 921
Query: 299 IFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQV 358
FD + EFD+F+EVR LFEGG+KLPTDI+S ISP+ +KEI RTDGE LK+PPP V
Sbjct: 922 FFDFD---KEFDNFEEVRCLFEGGVKLPTDIISAISPIAMVKEIFRTDGENFLKYPPPHV 1092
Query: 359 IRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNL 418
I+ KSAW TDEEF REMLAGVNP VI+ LQ FPP+S LD VYGDQ STITKEHLE N
Sbjct: 1093IQADKSAWMTDEEFAREMLAGVNPCVIQRLQVFPPESKLDPNVYGDQTSTITKEHLEINT 1272
Query: 419 GDITVEEALDGKRLFLLDYHDAFMPYLERINLN-AKAYATRTILFLQDDGTLKPLAIELS 477
+TVE+A+ +RLF+LDYHDAFM YL+ IN K+YATRTILFL+DDGTLKPLAIELS
Sbjct: 1273DGLTVEKAIQDERLFILDYHDAFMSYLKYINKPIPKSYATRTILFLKDDGTLKPLAIELS 1452
Query: 478 LPHSNGVQYGAESKVFLPAH-EGVESTIWMLAKAHVIVNDSCYHQLMSH 525
LPH +G +YGA SKV LP GV+ TIW LAKA+V+VND+C+HQLMSH
Sbjct: 1453LPHPDGEKYGAVSKVLLPPEGHGVQRTIWQLAKAYVVVNDACFHQLMSH 1599
Score = 75.5 bits (184), Expect(2) = 0.0
Identities = 47/83 (56%), Positives = 52/83 (62%)
Frame = +3
Query: 527 DGTIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH*D 586
D IHHS K* S+C S *A L *HN+YQ TCS +Q HYRA+I +RT *D
Sbjct: 1620 D*AIHHSYK*VSQCGSSYS*ASTASLS*HNEYQCTCSEFIDQWRWHYRASIFARTVRC*D 1799
Query: 587 FFCSLQELGFH*SSITS*SYQER 609
CSLQ LGF * S T *SYQER
Sbjct: 1800 VVCSLQGLGFS*PSTTC*SYQER 1868
>TC85192 homologue to PIR|T12142|T12142 lipoxygenase (EC 1.13.11.12) 1 -
fava bean, complete
Length = 2918
Score = 691 bits (1783), Expect(2) = 0.0
Identities = 338/518 (65%), Positives = 415/518 (79%), Gaps = 2/518 (0%)
Frame = +1
Query: 10 KIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATAFLGRNVSMQ 69
KIKGT+VLM KNVLD N++T K +I G G + DT T+FLGR++ +Q
Sbjct: 49 KIKGTLVLMQKNVLDINSLTDPTK---------IIDGALDGFGSIFDTLTSFLGRSICLQ 201
Query: 70 LISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPGAFYIRNYT 129
LIS+TK +G+G +GKE +L + + LPTLG +Q AFSI FEYD+NFGIPGAF I+N+
Sbjct: 202 LISSTKIGLTGEGKLGKEAYLKEAINNLPTLGDKQTAFSIEFEYDSNFGIPGAFKIKNFM 381
Query: 130 QAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPAPLNHFRE 189
EF LV +TL+DIPN G++ F CNSWVYN K+Y +RIFF N+ +LPS+TPAPL ++R+
Sbjct: 382 STEFLLVSLTLDDIPNVGTIHFVCNSWVYNAKNYLTDRIFFANNTFLPSETPAPLVYYRQ 561
Query: 190 EELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVRTGRK 249
EL+ LRGDGTGERKEWDR+YDYDVYNDLG+PD G + RP+LGGSS+ PYPRR RTGRK
Sbjct: 562 LELKTLRGDGTGERKEWDRVYDYDVYNDLGDPDKGQSYARPVLGGSSSLPYPRRGRTGRK 741
Query: 250 PTRKDLKSE-KPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFDLNFTPNE 308
PT D SE + ++Y+PRDE FGHLKSSDFL+YG+KS+SQDV+PL+QSV FD NFTPNE
Sbjct: 742 PTATDPNSESRSSSVYIPRDEAFGHLKSSDFLVYGLKSVSQDVIPLIQSV-FDTNFTPNE 918
Query: 309 FDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVSKSAWNT 368
FDSFD+V L+EGGIKLPTDILSQISPLP L EI RTDGEQ LKFP P+VI+VSKSAW T
Sbjct: 919 FDSFDDVLDLYEGGIKLPTDILSQISPLPVLSEIFRTDGEQFLKFPTPKVIQVSKSAWMT 1098
Query: 369 DEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDITVEEALD 428
D+EFGRE+LAGVNP +IR LQEFPPKS LD+ VYGD STIT+EH++ NL +TV+EA+
Sbjct: 1099DDEFGREILAGVNPGLIRSLQEFPPKSKLDSAVYGDHTSTITREHIQLNLDGLTVDEAIQ 1278
Query: 429 GKRLFLLDYHDAFMPYLERIN-LNAKAYATRTILFLQDDGTLKPLAIELSLPHSNGVQYG 487
K+LFLL++HD +PYL IN + KAYA+RT+LFL+ DGTLKPLAIELSLPH G Q+G
Sbjct: 1279NKKLFLLEHHDTIIPYLRLINSTSTKAYASRTVLFLKSDGTLKPLAIELSLPHPQGDQFG 1458
Query: 488 AESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
S V+LPA EGVE+T+W+LAKA+VIVNDSC+HQL+SH
Sbjct: 1459VVSNVYLPAIEGVEATVWLLAKAYVIVNDSCFHQLVSH 1572
Score = 38.1 bits (87), Expect(2) = 0.0
Identities = 30/80 (37%), Positives = 41/80 (50%)
Frame = +3
Query: 529 TIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH*DFF 588
TI +SNK T+ C S * L + +Y+ TC +QC +Y +IL + D F
Sbjct: 1599 TICYSNKQTT*CASSYL*TITSTLSRYYEYKCTC*IILSQCRWYY*KDILVGRVCYGDIF 1778
Query: 589 CSLQELGFH*SSITS*SYQE 608
SL LGF+ S *SYQ+
Sbjct: 1779 KSL*GLGFYRPSPPC*SYQK 1838
>TC85195 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (98%)
Length = 2903
Score = 661 bits (1705), Expect(2) = 0.0
Identities = 322/525 (61%), Positives = 404/525 (76%), Gaps = 2/525 (0%)
Frame = +3
Query: 3 GIFDKGQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATAFL 62
GI ++G+K+KGTVVLM KNVLD N +TSI V G IGG VG V+DTATAFL
Sbjct: 78 GIINRGEKLKGTVVLMQKNVLDVNELTSIKSNPVGGIIGGAIGGAFGAVGTVLDTATAFL 257
Query: 63 GRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPGA 122
GRNV+++LISAT D SGKG VGK+TFL + +PTLG +Q+A+S+ FE+ ++ GIPGA
Sbjct: 258 GRNVALKLISATSADGSGKGKVGKQTFLEGVITSIPTLGDKQNAYSVHFEWGSDMGIPGA 437
Query: 123 FYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPA 182
FYI N+ Q EFFLV +TLED+PN G++ F CNSWVYN K YK++RIFF N YLPS+TPA
Sbjct: 438 FYIENFLQHEFFLVSLTLEDVPNHGTINFVCNSWVYNDKKYKSDRIFFANKTYLPSETPA 617
Query: 183 PLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPR 242
PL ++R+EEL+ LRGDG GERKEW+RIYDYDVYNDLG PD L RP+LGGSST PYPR
Sbjct: 618 PLVYYRQEELKTLRGDGKGERKEWERIYDYDVYNDLGEPDSKPTLGRPVLGGSSTLPYPR 797
Query: 243 RVRTGRKPTRKDLKSE-KPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFD 301
R RTGRKP +KD KSE + G +Y+PRDE FGH KSSDFL + +KS SQ+++P L+SV+
Sbjct: 798 RGRTGRKPAKKDPKSESRSGTVYLPRDEAFGHTKSSDFLAFILKSASQNIIPSLKSVV-- 971
Query: 302 LNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRV 361
EF++F++VR L++GGIKLPT+ LS +SP+P E+ R+DG LKF PP+V++V
Sbjct: 972 ----SKEFNNFEDVRSLYDGGIKLPTNFLSNVSPIPLFTELFRSDGASTLKFSPPKVVQV 1139
Query: 362 SKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDI 421
SAW TDEEF REM+AGVNP++I+ L EFP KS LD+ YGD STITKE LE NLG I
Sbjct: 1140DHSAWMTDEEFAREMIAGVNPHIIKKLTEFPHKSKLDSQKYGDNTSTITKEQLEPNLGGI 1319
Query: 422 TVEEALDGKRLFLLDYHDAFMPYLERIN-LNAKAYATRTILFLQDDGTLKPLAIELSLPH 480
TVE+A+ +L++LD++D PYL +IN KAYA RTILFLQ+DGTLKPLAIELS PH
Sbjct: 1320TVEQAIQNNKLYILDHYDIVYPYLRKINATETKAYAARTILFLQNDGTLKPLAIELSKPH 1499
Query: 481 SNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
+ YG S+V+ PA EGVE++IW+LAKA+V+VNDSC+HQL+SH
Sbjct: 1500PDDDSYGPVSEVYFPASEGVEASIWLLAKAYVVVNDSCHHQLVSH 1634
Score = 57.0 bits (136), Expect(2) = 0.0
Identities = 36/86 (41%), Positives = 47/86 (53%)
Frame = +2
Query: 524 SHCDGTIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKF 583
S C +HHSNK T SP F LP*HN++++TC+ QC +YR NIL
Sbjct: 1646 SRCC*AVHHSNKQTP*YGSPCPQTFTSSLP*HNEHKFTCTECPCQC*GYYRINILVGRIC 1825
Query: 584 H*DFFCSLQELGFH*SSITS*SYQER 609
+ C + LGFH*+ T *S Q+R
Sbjct: 1826 FGNVCCCI*GLGFH*ARAT**STQKR 1903
>TC77404 homologue to GP|493730|emb|CAA55318.1| lipoxygenase {Pisum
sativum}, partial (85%)
Length = 2463
Score = 584 bits (1505), Expect(2) = e-177
Identities = 289/404 (71%), Positives = 333/404 (81%), Gaps = 2/404 (0%)
Frame = +3
Query: 124 YIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPAP 183
YI+NY Q EF+L +TLED+PN G+++F CNSWVYN K YK+ RIFF N +YLPS+TP+P
Sbjct: 3 YIKNYMQVEFYLKSLTLEDVPNHGTIRFVCNSWVYNAKLYKSPRIFFANKSYLPSETPSP 182
Query: 184 LNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRR 243
L +REEELQNLRGDGTGERK +RIYDYDVYNDLGNPD G++ RP+LGGSSTHPYPRR
Sbjct: 183 LVKYREEELQNLRGDGTGERKLHERIYDYDVYNDLGNPDHGESFARPVLGGSSTHPYPRR 362
Query: 244 VRTGRKPTRKDLKSEKPGA-IYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFDL 302
RTGR PTRKD SEKP IYVPRDENFGHLKSSDFL YGIKS+SQ+VLP +S FDL
Sbjct: 363 GRTGRYPTRKDPNSEKPATEIYVPRDENFGHLKSSDFLTYGIKSVSQNVLPAFESA-FDL 539
Query: 303 NFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVS 362
NFTP EFDSF +VR LFEGGIKLP D++S ISPLP +KE+ RTDGE LKFP P V++VS
Sbjct: 540 NFTPREFDSFQDVRDLFEGGIKLPLDVISTISPLPVIKELFRTDGENVLKFPTPHVVKVS 719
Query: 363 KSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDIT 422
KSAW TDEEF REMLAGVNP +IR L+EFPPKS LD TVYGDQ S IT + LE L T
Sbjct: 720 KSAWMTDEEFAREMLAGVNPCMIRGLKEFPPKSNLDPTVYGDQTSKITADALE--LDGST 893
Query: 423 VEEALDGKRLFLLDYHDAFMPYLERIN-LNAKAYATRTILFLQDDGTLKPLAIELSLPHS 481
V+EAL G RLF+LDYHD F+P+L RIN +AKAYATRTILFL++DG LKP+AIELSLPH
Sbjct: 894 VDEALAGGRLFILDYHDTFIPFLRRINETSAKAYATRTILFLKEDGNLKPVAIELSLPHP 1073
Query: 482 NGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
+G + G S+V LPA+EG E TIW+LAKA+V+VNDSCYHQLMSH
Sbjct: 1074DGDKSGVVSQVILPANEGGERTIWLLAKAYVVVNDSCYHQLMSH 1205
Score = 56.6 bits (135), Expect(2) = e-177
Identities = 37/80 (46%), Positives = 47/80 (58%)
Frame = +2
Query: 530 IHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH*DFFC 589
I + NK T+ C SP+ * FI L H+++Q TC SN H R NI + + F
Sbjct: 1235 IRYCNKSTA*CDSPNL*TFISSLSGHHEHQRTCKRISN*RQWHNRENIFAFKVCCGNVFS 1414
Query: 590 SLQELGFH*SSITS*SYQER 609
LQELGF *SS +S*SYQE+
Sbjct: 1415 GLQELGFP*SSPSS*SYQEK 1474
>TC85209 homologue to PIR|T06454|T06454 probable lipoxygenase (EC
1.13.11.12) - garden pea, partial (68%)
Length = 2002
Score = 453 bits (1166), Expect(2) = e-149
Identities = 222/261 (85%), Positives = 240/261 (91%)
Frame = +3
Query: 265 VPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFDLNFTPNEFDSFDEVRGLFEGGIK 324
VPRDENFGHLKSSDFL YGIKSLSQDVLPL +SVIFDLNFTPNEFDSFDEVR LFEGGI+
Sbjct: 3 VPRDENFGHLKSSDFLTYGIKSLSQDVLPLFKSVIFDLNFTPNEFDSFDEVRDLFEGGIE 182
Query: 325 LPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVSKSAWNTDEEFGREMLAGVNPNV 384
LPT ILS+ISPLP LKEI RTDGEQ LKFPPP VIRVSKSAW TDEEFGREM+AGVNP V
Sbjct: 183 LPTHILSKISPLPVLKEIFRTDGEQVLKFPPPHVIRVSKSAWMTDEEFGREMVAGVNPCV 362
Query: 385 IRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDITVEEALDGKRLFLLDYHDAFMPY 444
IRLLQEFPPKSTLD VYGDQ S + KEHLE NLG +TVE+AL+G+RLF+LDYHDAFMP+
Sbjct: 363 IRLLQEFPPKSTLDIAVYGDQTSILKKEHLEINLGGLTVEKALNGQRLFILDYHDAFMPF 542
Query: 445 LERINLNAKAYATRTILFLQDDGTLKPLAIELSLPHSNGVQYGAESKVFLPAHEGVESTI 504
LE+IN NAKAYATRTILFL+DDGTLKP+AIELSLPH NGV+YGAESKV LPA +GV+STI
Sbjct: 543 LEKINKNAKAYATRTILFLKDDGTLKPVAIELSLPHPNGVKYGAESKVILPADQGVDSTI 722
Query: 505 WMLAKAHVIVNDSCYHQLMSH 525
W+LAKAHVIVNDSCYHQLMSH
Sbjct: 723 WLLAKAHVIVNDSCYHQLMSH 785
Score = 94.7 bits (234), Expect(2) = e-149
Identities = 55/87 (63%), Positives = 60/87 (68%), Gaps = 1/87 (1%)
Frame = +2
Query: 526 CDG-TIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH 584
C G IHHSNK TS C+SP ** P P*HNKYQ TCS SN C + R NI + +F
Sbjct: 800 CSG*AIHHSNKQTS*CSSPY**VARSPFP*HNKYQRTCSKCSN*C*WNNRGNIFTWAEFC 979
Query: 585 *DFFCSLQELGFH*SSITS*SYQERIS 611
*D S+QELGFH*SSIT *SYQERIS
Sbjct: 980 *DVISSIQELGFH*SSITG*SYQERIS 1060
>CB892184 homologue to PIR|T06454|T06 probable lipoxygenase (EC 1.13.11.12) -
garden pea, partial (31%)
Length = 839
Score = 443 bits (1140), Expect = e-125
Identities = 227/331 (68%), Positives = 249/331 (74%), Gaps = 1/331 (0%)
Frame = +1
Query: 78 ASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPGAFYIRNYTQA-EFFLV 136
A+GKGLVGKE F GAFYIRNY QA EFFLV
Sbjct: 1 ANGKGLVGKEVFF------------------------------GAFYIRNYMQAHEFFLV 90
Query: 137 RVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPAPLNHFREEELQNLR 196
VT +DIPN SV+F CNSW+YNFK+YK +RIFFTND YLPSQTPAPL ++R+EELQNLR
Sbjct: 91 SVTFDDIPNHESVEFVCNSWIYNFKNYKKDRIFFTNDTYLPSQTPAPLVYYRQEELQNLR 270
Query: 197 GDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVRTGRKPTRKDLK 256
GDGTG+RKEW+R+YDYDVYNDLG+PD L RP+LGGSSTHPYPRRVR+GRKPT+KD K
Sbjct: 271 GDGTGQRKEWERVYDYDVYNDLGDPDEDVKLARPVLGGSSTHPYPRRVRSGRKPTKKDPK 450
Query: 257 SEKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFDLNFTPNEFDSFDEVR 316
SE+PG +YVPRDENFGHLKSSDFL YGIKSLSQDVLPL +SVIFDLNFTPNEFDSFDEVR
Sbjct: 451 SERPGVMYVPRDENFGHLKSSDFLTYGIKSLSQDVLPLFKSVIFDLNFTPNEFDSFDEVR 630
Query: 317 GLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVSKSAWNTDEEFGREM 376
LFEGGI+LPT ILS+ISPLP LKEI R TDEEFGREM
Sbjct: 631 DLFEGGIELPTHILSKISPLPVLKEIFR-----------------------TDEEFGREM 741
Query: 377 LAGVNPNVIRLLQEFPPKSTLDATVYGDQNS 407
+AGVNP VIRLLQEFPPKSTLD VYGDQ S
Sbjct: 742 VAGVNPCVIRLLQEFPPKSTLDIAVYGDQTS 834
>TC88044 similar to PIR|T06454|T06454 probable lipoxygenase (EC 1.13.11.12)
- garden pea, partial (28%)
Length = 804
Score = 398 bits (1023), Expect = e-111
Identities = 191/248 (77%), Positives = 219/248 (88%), Gaps = 3/248 (1%)
Frame = +2
Query: 1 MFGIFDK--GQKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTA 58
MFGIFD+ QKIKGTVVLMPKNVLDFNA+TS+ +GG LDAAGN++ S+VGG++D A
Sbjct: 59 MFGIFDRDHSQKIKGTVVLMPKNVLDFNAVTSVKQGGFLDAAGNVLDAAGSLVGGIIDGA 238
Query: 59 TAFLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFG 118
TAFLGRNVSM+LISATKTDA+GKGLVGKE FL KH+P LPTLGARQDAFSI F++DA+FG
Sbjct: 239 TAFLGRNVSMRLISATKTDANGKGLVGKEVFLEKHVPTLPTLGARQDAFSIHFDWDADFG 418
Query: 119 IPGAFYIRNYTQA-EFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLP 177
IPGAFYIRNY QA EFFLV VT +DIPN SV+F CNSW+YNFK+YK +RIFFTND YLP
Sbjct: 419 IPGAFYIRNYMQAHEFFLVSVTFDDIPNHESVEFVCNSWIYNFKNYKKDRIFFTNDTYLP 598
Query: 178 SQTPAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSST 237
SQTPAPL ++R+EELQNLRGDGTG+RKEW+R+YDYDVYNDLG+PD L RP+LGGSST
Sbjct: 599 SQTPAPLVYYRQEELQNLRGDGTGQRKEWERVYDYDVYNDLGDPDEDVKLARPVLGGSST 778
Query: 238 HPYPRRVR 245
HPYPRRVR
Sbjct: 779 HPYPRRVR 802
>TC85201 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11.12).
[Lentil] {Lens culinaris}, partial (49%)
Length = 1275
Score = 305 bits (782), Expect(2) = 9e-92
Identities = 145/225 (64%), Positives = 181/225 (80%), Gaps = 1/225 (0%)
Frame = +1
Query: 302 LNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRV 361
L F EF++F++VR L+EGGI+LPTDILS++SP+P KE+ RTDGE ALKFPPP+VI+V
Sbjct: 7 LQFHEPEFNTFEDVRSLYEGGIRLPTDILSELSPIPLFKELFRTDGEAALKFPPPKVIQV 186
Query: 362 SKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDI 421
SAW TDEEF REM+AGVNP++I+ L EFPPKS LD ++G+ STITKEHL+ N+ +
Sbjct: 187 DHSAWMTDEEFAREMIAGVNPHIIKKLLEFPPKSKLDTQLFGNNTSTITKEHLQPNMVGV 366
Query: 422 TVEEALDGKRLFLLDYHDAFMPYLERIN-LNAKAYATRTILFLQDDGTLKPLAIELSLPH 480
TVE+A+ +LF+LD+HD PYL +IN KAYATRTILFLQ+DGTLKPLAIELS PH
Sbjct: 367 TVEQAIQNNKLFILDHHDPLFPYLRKINATETKAYATRTILFLQNDGTLKPLAIELSRPH 546
Query: 481 SNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
G G S V+LPA EGVE++IW+LAKA+V+VNDSCYHQL+SH
Sbjct: 547 PQGDSLGPVSNVYLPASEGVEASIWLLAKAYVVVNDSCYHQLVSH 681
Score = 50.4 bits (119), Expect(2) = 9e-92
Identities = 34/80 (42%), Positives = 43/80 (53%)
Frame = +3
Query: 530 IHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH*DFFC 589
IHHSNK* S C SP * F LP*HN++++ + QC Y N L *+ C
Sbjct: 711 IHHSNK*AS*CGSPYS*TFTSTLP*HNEHKFPRTNYLGQCRGCYGINFLVGRLCC*NVCC 890
Query: 590 SLQELGFH*SSITS*SYQER 609
LQ L FH + + *S Q+R
Sbjct: 891 CLQGLEFHRARTS**SNQKR 950
>TC85619 similar to PIR|T11578|T11578 probable lipoxygenase (EC 1.13.11.12)
CPRD46 drought-inducible - cowpea, partial (75%)
Length = 2452
Score = 286 bits (731), Expect(2) = 2e-81
Identities = 163/366 (44%), Positives = 213/366 (57%), Gaps = 6/366 (1%)
Frame = +3
Query: 166 NRIFFTNDAYLPSQTPAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGD 225
N+ F +YLPS+TP L REEEL+ LRG+G GERK +DRIYDYDVYNDLG+PD
Sbjct: 96 NQFFSFEQSYLPSETPEGLIRLREEELKTLRGNGQGERKSFDRIYDYDVYNDLGDPDSDP 275
Query: 226 ALVRPILGGSSTHPYPRRVRTGRKPTRKDLKSEK-PGAIYVPRDENFGHLKSSDFLMYGI 284
L RP+LGG HPYPRR RTGR D SEK +YVPRDE+F LK F +
Sbjct: 276 ELKRPVLGGKE-HPYPRRCRTGRPRCDTDPLSEKRSNNVYVPRDESFSELKQLSFSANTL 452
Query: 285 KSLSQDVLPLLQSVIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQI--SPLPALKEI 342
+S +LP++++ + D N F F + LF G LP S LP L +
Sbjct: 453 QSAFHALLPVVKTAVIDKNL---GFPLFSAIDDLFNEGFNLPPQAEKGFLASVLPRLVRL 623
Query: 343 LRTDGEQALKFPPPQVIRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVY 402
+ L+F P + + W DEEFGR+ +AG+NP I+L+ E+P KS LD +Y
Sbjct: 624 VNEARNDILRFETPATMDKDRFFWFRDEEFGRQTIAGLNPCCIQLVTEWPLKSKLDPKIY 803
Query: 403 GDQNSTITKEHLETNL-GDITVEEALDGKRLFLLDYHDAFMPYLERINL--NAKAYATRT 459
G S IT E +E + G TVEEA+ K+LF+LDY+D F+P +E + Y +RT
Sbjct: 804 GPAESAITTEMIEQQIRGYATVEEAIKQKKLFILDYYDFFLPLVEEVRKLEGTTLYGSRT 983
Query: 460 ILFLQDDGTLKPLAIELSLPHSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCY 519
+ FL +D TL+P+AIEL+ P G E VF PA +W LAKAHV+ +DS Y
Sbjct: 984 LFFLNEDSTLRPVAIELARPPIGGKPEWRE--VFTPAWHSTGVWLWRLAKAHVLAHDSGY 1157
Query: 520 HQLMSH 525
HQL+SH
Sbjct: 1158HQLVSH 1175
Score = 35.8 bits (81), Expect(2) = 2e-81
Identities = 17/46 (36%), Positives = 24/46 (51%)
Frame = +2
Query: 524 SHCDGTIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQC 569
S C +HHSNK T+KC + + HN Q TC+ S+ +C
Sbjct: 1187 SLCHRALHHSNKQTTKCNASNLQTITSTFSIHNGNQCTCTRSTYKC 1324
>AW685636 similar to SP|P38414|LOX1 Lipoxygenase (EC 1.13.11.12). [Lentil]
{Lens culinaris}, partial (25%)
Length = 665
Score = 295 bits (754), Expect = 5e-80
Identities = 143/217 (65%), Positives = 175/217 (79%), Gaps = 1/217 (0%)
Frame = +3
Query: 184 LNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRR 243
L ++R+EEL+ LRGDGTGERKEW+RIYDYDVYNDLG PD L R ILGGSS PYPRR
Sbjct: 3 LVYYRQEELKTLRGDGTGERKEWERIYDYDVYNDLGEPDSKPQLARQILGGSSDFPYPRR 182
Query: 244 VRTGRKPTRKDLKSE-KPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFDL 302
RTGR+ T+ D KSE + +Y+PRDE+FGH KSSDFL+Y +KS SQ+V+P L+SV+ L
Sbjct: 183 GRTGRRKTKTDPKSETRSPTVYLPRDESFGHTKSSDFLVYILKSASQNVIPQLRSVV-TL 359
Query: 303 NFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVS 362
EF++F +VR L+EGGIKLPTD+LSQISP+P KE+ R+DGEQALKFPPP+VI+V
Sbjct: 360 QLNNPEFNTFQDVRSLYEGGIKLPTDVLSQISPIPLFKELFRSDGEQALKFPPPKVIQVD 539
Query: 363 KSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDA 399
SAW TDEEF REM+AGVNP++I+ L EFPPKS LD+
Sbjct: 540 HSAWQTDEEFAREMIAGVNPHIIKKLSEFPPKSKLDS 650
>AW691537 similar to PIR|S56655|S56 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (25%)
Length = 668
Score = 285 bits (729), Expect = 4e-77
Identities = 141/248 (56%), Positives = 175/248 (69%), Gaps = 1/248 (0%)
Frame = +2
Query: 118 GIPGAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLP 177
GIPGAFYI N+ Q EFFLV +TLED+PN G++ F CNSWVYN K YK++RIFF N
Sbjct: 2 GIPGAFYIENFLQHEFFLVSLTLEDVPNHGTINFVCNSWVYNDKKYKSDRIFFAN----- 166
Query: 178 SQTPAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSST 237
+ LRGDG GERKEW+RIYDYDVYNDLG PD L RP+LGGSST
Sbjct: 167 ---------------KTLRGDGKGERKEWERIYDYDVYNDLGEPDSKPTLGRPVLGGSST 301
Query: 238 HPYPRRVRTGRKPTRKDLKSE-KPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQ 296
PYPRR RTGRKP +KD KSE + G +Y+PRDE FGH KSSDFL + +KS SQ+++P L+
Sbjct: 302 LPYPRRGRTGRKPAKKDPKSESRSGTVYLPRDEAFGHTKSSDFLAFILKSASQNIIPSLK 481
Query: 297 SVIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPP 356
SV+ EF++F++VR L++GGIKLPT+ LS +SP+P E+ R+DG LKF PP
Sbjct: 482 SVV------SKEFNNFEDVRSLYDGGIKLPTNFLSNVSPIPLFTELFRSDGASTLKFSPP 643
Query: 357 QVIRVSKS 364
+V +V S
Sbjct: 644 KVGQVDHS 667
>TC85183 homologue to SP|P09918|LOX3_PEA Seed lipoxygenase-3 (EC
1.13.11.12). [Garden pea] {Pisum sativum}, partial (57%)
Length = 1623
Score = 227 bits (579), Expect(2) = 5e-73
Identities = 114/157 (72%), Positives = 129/157 (81%), Gaps = 1/157 (0%)
Frame = +3
Query: 370 EEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDITVEEALDG 429
EEF REMLAGVNPNVI LQEFPP+S LD+ VYGD S ITKEHLE NL +TVEEA+
Sbjct: 3 EEFAREMLAGVNPNVICCLQEFPPRSKLDSQVYGDHTSKITKEHLEPNLEGLTVEEAIQN 182
Query: 430 KRLFLLDYHDAFMPYLERINLN-AKAYATRTILFLQDDGTLKPLAIELSLPHSNGVQYGA 488
K+LFLLD+HD+ MPYL RIN KAYATRTILFL D TLKPLAIELSLPH +G ++GA
Sbjct: 183 KKLFLLDHHDSIMPYLRRINSTPTKAYATRTILFLSSDKTLKPLAIELSLPHPDGDEHGA 362
Query: 489 ESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
S V+ PA EGVESTIW+LAKA+V+VNDSCYHQL+SH
Sbjct: 363 VSHVYQPALEGVESTIWLLAKAYVVVNDSCYHQLVSH 473
Score = 66.2 bits (160), Expect(2) = 5e-73
Identities = 39/81 (48%), Positives = 50/81 (61%)
Frame = +2
Query: 529 TIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH*DFF 588
TI HSN S+ +SP * F+ L HN+Y ++CST Q WRH+R NI F+ + F
Sbjct: 500 TICHSN*QASELSSPHL*TFVSSLSRHNEY*FSCSTIPGQRWRHHRKNIFVGKIFYGNVF 679
Query: 589 CSLQELGFH*SSITS*SYQER 609
SLQEL F SSI *SY++R
Sbjct: 680 *SLQELDFARSSIAC*SYKKR 742
>TC87113 weakly similar to GP|8649004|emb|CAB94852.1 lipoxygenase {Prunus
dulcis}, partial (32%)
Length = 1032
Score = 263 bits (673), Expect = 1e-70
Identities = 151/325 (46%), Positives = 192/325 (58%), Gaps = 9/325 (2%)
Frame = +1
Query: 11 IKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATAFLGRNVSMQL 70
+KG VVLM K++LDF+ I S V+D FLG+ VS+QL
Sbjct: 118 VKGKVVLMKKSLLDFHDIKS----------------------NVLDRIHEFLGKGVSLQL 231
Query: 71 ISATKTD-ASG-KGLVGKETFLSKHLPQLPTLGARQDA-FSIFFEYD-ANFGIPGAFYIR 126
ISAT D A G KG GK L + + + +L D FS+ F++D G+PGAF IR
Sbjct: 232 ISATAPDPAKGLKGKHGKVACLERWMSSISSLTTATDTEFSVTFDWDHEKMGVPGAFLIR 411
Query: 127 NYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPAPLNH 186
N ++F+L VTL+DIP G + F CNSWVY Y ++R+FF N AYLP TP L
Sbjct: 412 NNHHSQFYLKTVTLDDIPGHGPITFVCNSWVYPTHRYTHDRVFFANKAYLPCDTPEALRK 591
Query: 187 FREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVRT 246
REEEL LRG G G+ EWDR+YDY YNDLG PD G RP++GGS PYPRR RT
Sbjct: 592 LREEELGTLRGKGIGKLNEWDRVYDYACYNDLGTPDNGPDYARPVIGGSQKFPYPRRGRT 771
Query: 247 GRKPTRKDLKSEK-----PGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFD 301
R T+ D K+E IYVPRDE FGH+K SDFL Y +KS++Q +LP L+S+
Sbjct: 772 SRPHTKTDPKTESRLHLLNLNIYVPRDEQFGHVKFSDFLAYSLKSVTQVLLPELRSL--- 942
Query: 302 LNFTPNEFDSFDEVRGLFEGGIKLP 326
+ T NEFD+F +V ++EG LP
Sbjct: 943 CDKTINEFDTFQDVLDIYEGSFNLP 1017
>TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (75%)
Length = 2646
Score = 168 bits (425), Expect(2) = 7e-69
Identities = 89/186 (47%), Positives = 116/186 (61%), Gaps = 1/186 (0%)
Frame = +1
Query: 15 VVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATAFLGRNVSMQLISAT 74
VVLM KN L++ +IGG T + L +V +LISA+
Sbjct: 61 VVLMKKNALNYK----------------VIGGNTVVENEGDTVRPTLLDTSVGFKLISAS 192
Query: 75 KTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDAN-FGIPGAFYIRNYTQAEF 133
K DA+GKG VGKETF+ + +P LG Q+AFSI FE+D N GIPGAFY++N+TQ E
Sbjct: 193 KADATGKGKVGKETFMDGFVTSIPNLGDIQNAFSIHFEWDPNHMGIPGAFYVKNFTQDEI 372
Query: 134 FLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPAPLNHFREEELQ 193
FLV +TLED+ + + F CNSW+YN + Y+ RIFF N AYL +TPAPL ++R+EEL
Sbjct: 373 FLVSLTLEDVESHETTNFICNSWIYNAEKYQTERIFFANKAYLLRETPAPLLYYRQEELN 552
Query: 194 NLRGDG 199
LRGDG
Sbjct: 553 TLRGDG 570
Score = 167 bits (422), Expect(2) = 1e-51
Identities = 87/177 (49%), Positives = 120/177 (67%), Gaps = 3/177 (1%)
Frame = +1
Query: 352 KFPPPQVIRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITK 411
KF P VI+ ++AW TDEEF REM+AGVNPNVI+ + +TI K
Sbjct: 946 KFSTPLVIQEDRTAWMTDEEFAREMIAGVNPNVIKKV---------------GNTTTINK 1080
Query: 412 EHLETNLGD-ITVEEALDGKRLFLLDYHDAFMPYLERINLN--AKAYATRTILFLQDDGT 468
+H E + D ++VE+ + RL+++DY DA +PYL +IN N AKAYA+ T+L LQDDGT
Sbjct: 1081 KHFEPYMQDGVSVEQTIKDLRLYVVDYQDAILPYLRKINANGAAKAYASTTLLSLQDDGT 1260
Query: 469 LKPLAIELSLPHSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
LKP++IEL +PH +G G + + PA EGV+++IW LAKA+ +VND+CYHQL+SH
Sbjct: 1261 LKPISIELHVPHPDGD--GIVTTSYTPATEGVDASIWRLAKAYAVVNDACYHQLISH 1425
Score = 111 bits (278), Expect(2) = 7e-69
Identities = 58/98 (59%), Positives = 68/98 (69%), Gaps = 1/98 (1%)
Frame = +3
Query: 200 TGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVRTGRKPTRKDLKSE- 258
TGERKEWDRIYDYDVYNDLG PD L RP+LGGS+ PYPRR RTGRK K ++E
Sbjct: 570 TGERKEWDRIYDYDVYNDLGQPDQNPCLYRPVLGGSTALPYPRRGRTGRKHLEKYPETES 749
Query: 259 KPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQ 296
+ Y+PRDE KSSDF++ IKS+SQ P L+
Sbjct: 750 RSNYFYIPRDELIIPPKSSDFVVNTIKSISQYETPQLK 863
Score = 55.5 bits (132), Expect(2) = 1e-51
Identities = 41/95 (43%), Positives = 51/95 (53%), Gaps = 2/95 (2%)
Frame = +3
Query: 517 SCYHQLMSHCDG--TIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYR 574
S Y L++H IHHSNK* S C SP * L *HN+++ C+ +Q +YR
Sbjct: 1410 STYQPLVAHSCFC*AIHHSNK*AS*CGSPYS*TITSTLS*HNEHKCQCARCLDQSRGYYR 1589
Query: 575 ANILSRTKFH*DFFCSLQELGFH*SSITS*SYQER 609
NIL F+ F LQ LGF SS T *S Q+R
Sbjct: 1590 VNILVWKLFNGIVF*CLQGLGFPGSSTT**SNQKR 1694
>TC85173 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11.12).
[Lentil] {Lens culinaris}, partial (22%)
Length = 643
Score = 245 bits (625), Expect = 4e-65
Identities = 118/197 (59%), Positives = 151/197 (75%), Gaps = 1/197 (0%)
Frame = +1
Query: 3 GIFDKGQKIKGTVVLMPKNVLDFNAITSI-GKGGVLDAAGNLIGGVTSIVGGVVDTATAF 61
G+FD+GQK+KGTVVLM KN LD NA+T+ G++ A ++G V G ++DTAT+F
Sbjct: 61 GLFDRGQKLKGTVVLMQKNTLDINALTAAKSPTGIIGGAFGIVGDVA---GNILDTATSF 231
Query: 62 LGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPG 121
LGR+++++LISAT DA+GKG VGKETFL L +PTLG +Q+AFSI FE+D+N GIPG
Sbjct: 232 LGRSIALRLISATSADAAGKGKVGKETFLEGLLTSIPTLGDKQNAFSIHFEWDSNMGIPG 411
Query: 122 AFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTP 181
AFYI N+ Q EFFLV +TLED+PN GS+ F CNSW+YN K YK +RIFF N YLPS+TP
Sbjct: 412 AFYIDNFMQGEFFLVSLTLEDVPNHGSIHFVCNSWIYNSKKYKTDRIFFANKTYLPSETP 591
Query: 182 APLNHFREEELQNLRGD 198
APL ++R+ EL LRGD
Sbjct: 592 APLVYYRQ*ELNTLRGD 642
>TC85194 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (14%)
Length = 887
Score = 243 bits (621), Expect = 1e-64
Identities = 122/217 (56%), Positives = 157/217 (72%), Gaps = 10/217 (4%)
Frame = +3
Query: 63 GRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDAN---FGI 119
GR+V+++LIS + D +GKG VG+ T+L + T Q F+I FE+D
Sbjct: 141 GRSVALRLISDSTVDGNGKGKVGERTYLGSLIASDET----QFHFNITFEWDTEKLGAPS 308
Query: 120 PGAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQ 179
PGAF+I N+ Q+E FLV +TL D+PN G++ F+CNSW+YN K+Y + RIFFTN YL S+
Sbjct: 309 PGAFFIENFNQSELFLVSLTLNDVPNHGTINFNCNSWIYNAKNYHSERIFFTNKTYLLSE 488
Query: 180 TPAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSST-- 237
TPAPL ++R+EEL+ LRGDGTGERKEWDRIYDYDVYNDLG PD L RP+LGGSST
Sbjct: 489 TPAPLVYYRQEELKTLRGDGTGERKEWDRIYDYDVYNDLGEPDKEAGLGRPVLGGSSTLE 668
Query: 238 ----HPYPRRVRTGRKPTRKDLKSE-KPGAIYVPRDE 269
PYPRRVR+GR+P++KD K+E + G IY+PRDE
Sbjct: 669 GSDGFPYPRRVRSGREPSKKDPKTESRTGRIYIPRDE 779
>TC85210 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (18%)
Length = 863
Score = 211 bits (536), Expect = 9e-55
Identities = 111/224 (49%), Positives = 144/224 (63%), Gaps = 1/224 (0%)
Frame = +3
Query: 205 EWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVRTGRKPTRKDLKSE-KPGAI 263
EW+RIYDYDVYNDLG PD L R ILGGSS PYPRR RTGR+P +KD KSE + G +
Sbjct: 3 EWERIYDYDVYNDLGEPDSKPQLARQILGGSSNFPYPRRGRTGREPAKKDPKSESRNGFV 182
Query: 264 YVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFDLNFTPNEFDSFDEVRGLFEGGI 323
Y+PRDE+F H KSS+FL +KS SQD + + + N+ P EFD+F++V ++ +
Sbjct: 183 YIPRDESFDHKKSSEFLDNLLKSASQD---FISEIEIECNYKP-EFDTFNDVHAFYDEEV 350
Query: 324 KLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVSKSAWNTDEEFGREMLAGVNPN 383
L FP P+VI+V++S W TDEEF REM+AGVNP+
Sbjct: 351 P-------------------------GLTFPRPEVIQVNQSGWMTDEEFTREMIAGVNPH 455
Query: 384 VIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDITVEEAL 427
+I+ LQEFPPKS L++ YGD STITKE L+ N+ +TVEE +
Sbjct: 456 IIKRLQEFPPKSKLNSQDYGDNTSTITKEQLQLNMDGVTVEEVM 587
>TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Corylus
avellana}, partial (54%)
Length = 1763
Score = 190 bits (483), Expect(2) = 3e-52
Identities = 90/136 (66%), Positives = 110/136 (80%), Gaps = 1/136 (0%)
Frame = +3
Query: 391 FPPKSTLDATVYGDQNSTITKEHLETNLGDITVEEALDGKRLFLLDYHDAFMPYLERIN- 449
FPP S LD +VYGDQNS+I +H+E +L +T++EAL+ +L++LD+HDA MPYL RIN
Sbjct: 12 FPPASKLDPSVYGDQNSSIQAKHIENSLDGLTIDEALESDKLYILDHHDALMPYLSRINS 191
Query: 450 LNAKAYATRTILFLQDDGTLKPLAIELSLPHSNGVQYGAESKVFLPAHEGVESTIWMLAK 509
N K YATRT+LFLQDDGTLKPLAIELSLPH G Q+GA SKVF P+HEGV +T+W LAK
Sbjct: 192 TNTKTYATRTLLFLQDDGTLKPLAIELSLPHPQGEQHGAVSKVFTPSHEGVAATVWQLAK 371
Query: 510 AHVIVNDSCYHQLMSH 525
A+ VNDS YHQL+SH
Sbjct: 372 AYAAVNDSGYHQLVSH 419
Score = 33.9 bits (76), Expect(2) = 3e-52
Identities = 29/85 (34%), Positives = 42/85 (49%)
Frame = +2
Query: 527 DGTIHHSNK*TSKCTSPD**AFIPPLP*HNKYQWTCSTSSNQCWRHYRANILSRTKFH*D 586
D TI++S K T++ +S + *A +N Y+ + Q WR R N +
Sbjct: 440 D*TIYNSYKQTTESSSSNS*ALEATFQGYNAYKCLS*AYTYQRWRGTRENSFPW*ICIGN 619
Query: 587 FFCSLQELGFH*SSITS*SYQERIS 611
C + +LG H*+S T *S QER S
Sbjct: 620 VSCCI*KLGIH*TSTTC*SAQERHS 694
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.142 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,638,450
Number of Sequences: 36976
Number of extensions: 279218
Number of successful extensions: 1847
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 1739
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1778
length of query: 611
length of database: 9,014,727
effective HSP length: 102
effective length of query: 509
effective length of database: 5,243,175
effective search space: 2668776075
effective search space used: 2668776075
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140032.2


