

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140030.7 - phase: 0
(271 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
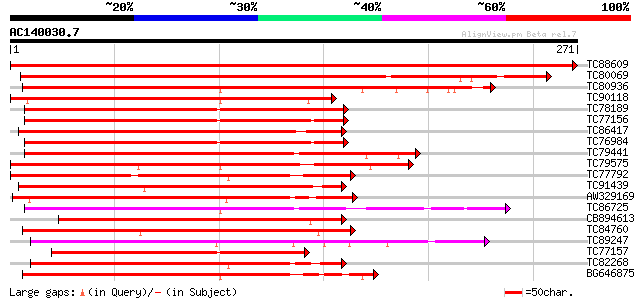
Score E
Sequences producing significant alignments: (bits) Value
TC88609 similar to GP|15148914|gb|AAK84884.1 NAC domain protein ... 568 e-162
TC80069 similar to GP|15148914|gb|AAK84884.1 NAC domain protein ... 370 e-103
TC80936 similar to GP|15795116|dbj|BAB02380. jasmonic acid regul... 239 1e-63
TC90118 homologue to GP|15795116|dbj|BAB02380. jasmonic acid reg... 223 5e-59
TC78189 homologue to GP|14485513|emb|CAC42087. putative NAC doma... 223 5e-59
TC77156 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-li... 221 2e-58
TC86417 similar to GP|16226943|gb|AAL16305.1 AT4g27410/F27G19_10... 218 1e-57
TC76984 similar to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like... 216 6e-57
TC79441 similar to GP|9757865|dbj|BAB08499.1 NAM (no apical meri... 189 7e-49
TC79575 similar to GP|7715611|gb|AAF68129.1| F20B17.1 {Arabidops... 187 4e-48
TC77792 similar to GP|1418990|emb|CAA99760.1 unknown {Lycopersic... 186 8e-48
TC91439 similar to PIR|D96683|D96683 hypothetical protein F12P19... 182 1e-46
AW329169 similar to PIR|E84636|E84 NAM (no apical meristem)-like... 181 2e-46
TC86725 similar to GP|9758909|dbj|BAB09485.1 NAM (no apical meri... 180 6e-46
CB894613 similar to GP|16226943|gb AT4g27410/F27G19_10 {Arabidop... 177 5e-45
TC84760 similar to GP|8567777|gb|AAF76349.1| unknown protein {Ar... 170 5e-43
TC89247 similar to GP|20303588|gb|AAM19015.1 putative NAM (no ap... 167 5e-42
TC77157 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-li... 167 5e-42
TC82268 homologue to GP|20197337|gb|AAC78526.2 NAM (no apical me... 165 2e-41
BG646875 similar to GP|21593065|gb NAM / CUC2-like protein {Arab... 164 4e-41
>TC88609 similar to GP|15148914|gb|AAK84884.1 NAC domain protein NAC2
{Phaseolus vulgaris}, partial (71%)
Length = 1076
Score = 568 bits (1463), Expect = e-162
Identities = 270/271 (99%), Positives = 270/271 (99%)
Frame = +1
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYK DPWELPDKSEF
Sbjct: 7 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKCDPWELPDKSEF 186
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP
Sbjct: 187 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 366
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN 180
KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN
Sbjct: 367 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN 546
Query: 181 DSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDGSTLDFQINNSNIGIDPYVKPQ 240
DSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDGSTLDFQINNSNIGIDPYVKPQ
Sbjct: 547 DSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDGSTLDFQINNSNIGIDPYVKPQ 726
Query: 241 PVEMTNHYEADSHSSITNQPIFVKQMHNYLA 271
PVEMTNHYEADSHSSITNQPIFVKQMHNYLA
Sbjct: 727 PVEMTNHYEADSHSSITNQPIFVKQMHNYLA 819
>TC80069 similar to GP|15148914|gb|AAK84884.1 NAC domain protein NAC2
{Phaseolus vulgaris}, partial (63%)
Length = 1058
Score = 370 bits (950), Expect = e-103
Identities = 182/260 (70%), Positives = 208/260 (80%), Gaps = 6/260 (2%)
Frame = +2
Query: 6 SSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEW 65
S ELPPGFRFHPTDEELIV+YLCNQATS+PCPASIIPEVDIYKFDPWELPDKS F ENEW
Sbjct: 128 SMELPPGFRFHPTDEELIVYYLCNQATSQPCPASIIPEVDIYKFDPWELPDKSGFGENEW 307
Query: 66 YFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKT 125
YFF+PRERKYPNGVRPNRAT+SGYWKATGTDK+I SGSK IGVKK+LVFYKGRPPKG+KT
Sbjct: 308 YFFTPRERKYPNGVRPNRATVSGYWKATGTDKSIFSGSKHIGVKKALVFYKGRPPKGIKT 487
Query: 126 DWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFNDSIIT 185
DWIMHEYRLIGS++Q ++ IGSMRLDDWVLCRIYKKK++ K+L+ EDY T+Q N +
Sbjct: 488 DWIMHEYRLIGSRRQANRQIGSMRLDDWVLCRIYKKKNITKSLETNEDYPTNQIN--MTQ 661
Query: 186 NNDDGELEMMNLTRSCSLTYLLDMNYFGP---ILSDG---STLDFQINNSNIGIDPYVKP 239
NDD E E++ R+CSLT LLDM+Y GP ILSDG ST +FQIN ++ G + P
Sbjct: 662 TNDDNEQELVKFPRTCSLTNLLDMDYMGPISQILSDGSYNSTFEFQINTAHGGT---IDP 832
Query: 240 QPVEMTNHYEADSHSSITNQ 259
Y +SS NQ
Sbjct: 833 TYAAGLEKYNVKQNSSFGNQ 892
>TC80936 similar to GP|15795116|dbj|BAB02380. jasmonic acid regulatory
protein-like {Arabidopsis thaliana}, partial (38%)
Length = 1254
Score = 239 bits (609), Expect = 1e-63
Identities = 132/249 (53%), Positives = 157/249 (63%), Gaps = 23/249 (9%)
Frame = +2
Query: 7 SELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWY 66
+ LPPGFRFHPTD ELI+HYL + S P P SII EVDIYK DPW+LP K+ F E EWY
Sbjct: 50 TNLPPGFRFHPTDAELILHYLRKKIASIPLPVSIIAEVDIYKLDPWDLPAKASFGEKEWY 229
Query: 67 FFSPRERKYPNGVRPNRATLSGYWKATGTDKAI---------KSGSKQIGVKKSLVFYKG 117
FFSPR+RKYPNG RPNRA SGYWKATGTDK I +S IGVKK+LVFYKG
Sbjct: 230 FFSPRDRKYPNGARPNRAAASGYWKATGTDKTIVASLPCGGGRSQENIIGVKKALVFYKG 409
Query: 118 RPPKGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKT--------LQ 169
+PPKG+KT+WIMHEYRL+ + K SMRLDDWVLCRIYKK T +
Sbjct: 410 KPPKGIKTNWIMHEYRLVDNNKPIKLKDSSMRLDDWVLCRIYKKSKCALTSTESSETMVG 589
Query: 170 QKEDYSTHQFNDSI--ITNNDDGELEMMNLT-RSCSLTYLLD-MNY--FGPILSDGSTLD 223
+ E QF +++ IT N L+ ++ +S S + LLD M+Y LS+ S
Sbjct: 590 EVEHAEETQFQETLFPITKNTTSPLQNTLMSQKSVSFSNLLDAMDYSMLSSFLSENSN-- 763
Query: 224 FQINNSNIG 232
N S IG
Sbjct: 764 ---NPSGIG 781
>TC90118 homologue to GP|15795116|dbj|BAB02380. jasmonic acid regulatory
protein-like {Arabidopsis thaliana}, partial (43%)
Length = 612
Score = 223 bits (569), Expect = 5e-59
Identities = 106/171 (61%), Positives = 126/171 (72%), Gaps = 15/171 (8%)
Frame = +3
Query: 1 MESSASS------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWEL 54
MES+ SS LPPGFRFHPTDEEL+VHYL +A S P P +II EVD+YKFDPWEL
Sbjct: 99 MESTDSSTGSQQPNLPPGFRFHPTDEELVVHYLKKKAASAPLPVAIIAEVDLYKFDPWEL 278
Query: 55 PDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKKSL 112
P K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + G++++GVKK+L
Sbjct: 279 PAKATFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVLTSGGTQKVGVKKAL 458
Query: 113 VFYKGRPPKGVKTDWIMHEYRLIGSQKQT-------SKHIGSMRLDDWVLC 156
VFY G+PP+G+KT+WIMHEYRL ++ S+RLDDWVLC
Sbjct: 459 VFYGGKPPRGIKTNWIMHEYRLADNKPNNRPPGCDWGNKKNSLRLDDWVLC 611
>TC78189 homologue to GP|14485513|emb|CAC42087. putative NAC domain protein
{Solanum tuberosum}, partial (56%)
Length = 1556
Score = 223 bits (569), Expect = 5e-59
Identities = 99/155 (63%), Positives = 119/155 (75%)
Frame = +2
Query: 8 ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYF 67
ELPPGFRFHPTD+EL+ HYLC + S P II E+D+YKFDPW LP+ + + E EWYF
Sbjct: 122 ELPPGFRFHPTDDELVNHYLCRKCASLPIAVPIIKEIDLYKFDPWHLPEMALYGEKEWYF 301
Query: 68 FSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
FSPR+RKYPNG RPNRA +GYWKATG DK I K +G+KK+LVFY G+ PKG+KT+W
Sbjct: 302 FSPRDRKYPNGSRPNRAAGTGYWKATGADKPI-GKPKALGIKKALVFYAGKAPKGIKTNW 478
Query: 128 IMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK 162
IMHEYRL + SK+ ++RLDDWVLCRIY KK
Sbjct: 479 IMHEYRLANVDRSASKNNHNLRLDDWVLCRIYNKK 583
>TC77156 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like protein
{Arabidopsis thaliana}, partial (66%)
Length = 1096
Score = 221 bits (564), Expect = 2e-58
Identities = 100/155 (64%), Positives = 119/155 (76%)
Frame = +3
Query: 8 ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYF 67
+LPPGFRFHPTDEEL++HYLC + TS+P II E+D+YK+DPW+LP + + E EWYF
Sbjct: 255 QLPPGFRFHPTDEELVMHYLCRKCTSQPIAVPIIAEIDLYKYDPWDLPGMATYGEKEWYF 434
Query: 68 FSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
FSPR+RKYPNG RPNRA SGYWKATG DK I K +G+KK+LVFY G+ PKG KT+W
Sbjct: 435 FSPRDRKYPNGSRPNRAAGSGYWKATGADKPI-GHPKPVGIKKALVFYAGKAPKGDKTNW 611
Query: 128 IMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK 162
IMHEYRL + K S+RLDDWVLCRIY KK
Sbjct: 612 IMHEYRLADVDRSIRKK-NSLRLDDWVLCRIYNKK 713
>TC86417 similar to GP|16226943|gb|AAL16305.1 AT4g27410/F27G19_10
{Arabidopsis thaliana}, partial (67%)
Length = 1628
Score = 218 bits (556), Expect = 1e-57
Identities = 99/157 (63%), Positives = 118/157 (75%)
Frame = +2
Query: 5 ASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENE 64
A LPPGFRF+PTDEEL+V YLC + II E+D+YKFDPW LP K+ F E E
Sbjct: 164 AQLSLPPGFRFYPTDEELLVQYLCRKVAGHHFSLQIIAEIDLYKFDPWILPSKAIFGEKE 343
Query: 65 WYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVK 124
WYFFSPR+RKYPNG RPNR SGYWKATGTDK I + +++G+KK+LVFY G+ PKG K
Sbjct: 344 WYFFSPRDRKYPNGTRPNRVAGSGYWKATGTDKIITNEGRKVGIKKALVFYVGKAPKGTK 523
Query: 125 TDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKK 161
T+WIMHEYRL+ S S++ G +LDDWVLCRIYKK
Sbjct: 524 TNWIMHEYRLLDS----SRNNGGTKLDDWVLCRIYKK 622
>TC76984 similar to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like protein
{Arabidopsis thaliana}, partial (71%)
Length = 1450
Score = 216 bits (551), Expect = 6e-57
Identities = 98/155 (63%), Positives = 115/155 (73%)
Frame = +2
Query: 8 ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYF 67
ELPPGFRFHPTDEEL+ HYLC + + ++ EVD+YKFDPW+LP+ E EWYF
Sbjct: 152 ELPPGFRFHPTDEELVNHYLCRKCAGQSISVPVVKEVDLYKFDPWQLPEMGYHSEKEWYF 331
Query: 68 FSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
FSPR+RKYPNG RPNRA SGYWKATG DK I K +G+KK+LVFY G+ PKGVKT+W
Sbjct: 332 FSPRDRKYPNGSRPNRAAGSGYWKATGADKPI-GKLKPMGIKKALVFYAGKAPKGVKTNW 508
Query: 128 IMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK 162
IMHEYRL + K ++RLDDWVLCRIY KK
Sbjct: 509 IMHEYRLANVDRSAGKK-NNLRLDDWVLCRIYNKK 610
>TC79441 similar to GP|9757865|dbj|BAB08499.1 NAM (no apical meristem)-like
protein {Arabidopsis thaliana}, partial (61%)
Length = 1247
Score = 189 bits (481), Expect = 7e-49
Identities = 92/198 (46%), Positives = 129/198 (64%), Gaps = 9/198 (4%)
Frame = +3
Query: 8 ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYF 67
+LPPGFRFHPTDEELI HYL + A I +VD+ K +PW+LP K++ E EWYF
Sbjct: 186 DLPPGFRFHPTDEELISHYLYKKVIDSNFSARAIGDVDLNKSEPWDLPFKAKMGEKEWYF 365
Query: 68 FSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
F R+RKYP G+R NRAT +GYWKATG DK I G +G+KK+LVFYKGR PKG K++W
Sbjct: 366 FCVRDRKYPTGLRTNRATEAGYWKATGKDKEIFKGKSLVGMKKTLVFYKGRAPKGEKSNW 545
Query: 128 IMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQ----QKEDYSTHQFNDSI 183
+MHEYRL G K + ++ ++WV+CR+++K GK + D ++F S+
Sbjct: 546 VMHEYRLEG--KFSVHNLPKAAKNEWVICRVFQKSSAGKKTHISGIMRVDSLGNEFGCSV 719
Query: 184 I-----TNNDDGELEMMN 196
+ ++N G+++ +N
Sbjct: 720 LPPLTDSSNSIGKIKQLN 773
>TC79575 similar to GP|7715611|gb|AAF68129.1| F20B17.1 {Arabidopsis
thaliana}, partial (58%)
Length = 1324
Score = 187 bits (475), Expect = 4e-48
Identities = 93/198 (46%), Positives = 127/198 (63%), Gaps = 5/198 (2%)
Frame = +2
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M+ S +PPGFRFHPTDEEL+ +YL + + + +I EVD+ + +PW+L DK
Sbjct: 104 MQGSGELTVPPGFRFHPTDEELLYYYLRKKVSYEAIDLDVIREVDLNRLEPWDLKDKCRI 283
Query: 61 ---EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYK 116
+NEWYFFS +++KYP G R NRAT +G+WKATG DKAI + SK+IG++K+LVFY
Sbjct: 284 GSGPQNEWYFFSHKDKKYPTGTRTNRATTAGFWKATGRDKAIYHTNSKRIGMRKTLVFYT 463
Query: 117 GRPPKGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQK-EDYS 175
GR P G KTDWIMHEYRL ++ D WV+CR++KKK+ ++ QQ+ E+
Sbjct: 464 GRAPHGHKTDWIMHEYRLDDDDN------AEVQEDGWVVCRVFKKKNTNRSYQQEIEEED 625
Query: 176 THQFNDSIITNNDDGELE 193
H N TN + LE
Sbjct: 626 HHMINHMRSTNGPNHFLE 679
>TC77792 similar to GP|1418990|emb|CAA99760.1 unknown {Lycopersicon
esculentum}, partial (52%)
Length = 1022
Score = 186 bits (472), Expect = 8e-48
Identities = 94/167 (56%), Positives = 113/167 (67%), Gaps = 2/167 (1%)
Frame = +1
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
++ + LPPGFRFHPTDEEL+V YL + S P PASIIPEVD+ K DPW+LP
Sbjct: 412 VKKNGEVRLPPGFRFHPTDEELVVQYLKRKVFSFPLPASIIPEVDLCKSDPWDLPGDM-- 585
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGS--KQIGVKKSLVFYKGR 118
E E YFFS +E KYPNG R NRAT SGYWKATG DK I + + G+KK+LVFY+G+
Sbjct: 586 -EQERYFFSTKEAKYPNGNRSNRATNSGYWKATGLDKQIMNSKTHEVAGMKKTLVFYRGK 762
Query: 119 PPKGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMG 165
PP G +TDWIMHEYRL TS H ++WVLCRI+ K+ G
Sbjct: 763 PPHGSRTDWIMHEYRL------TSSHSNPPLNENWVLCRIFLKRRSG 885
>TC91439 similar to PIR|D96683|D96683 hypothetical protein F12P19.8
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1154
Score = 182 bits (462), Expect = 1e-46
Identities = 86/159 (54%), Positives = 110/159 (69%), Gaps = 2/159 (1%)
Frame = +3
Query: 5 ASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEEN- 63
A LPPGFRFHPTDEEL+ +YL + II EVD+YK +PW+LP KS N
Sbjct: 39 APVSLPPGFRFHPTDEELVAYYLNRKINGHKIELEIIAEVDLYKCEPWDLPGKSLLPGND 218
Query: 64 -EWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKG 122
EWYFFSPR+RKYPNG R NRAT SGYWKATG D+ + S ++ +GVKK+LV+Y+GR P G
Sbjct: 219 MEWYFFSPRDRKYPNGSRTNRATKSGYWKATGKDRKVNSQARAVGVKKTLVYYRGRAPHG 398
Query: 123 VKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKK 161
+T+W+MHEYRL + +T+ + D + LCR+ KK
Sbjct: 399 SRTNWVMHEYRLDERECETTPSL----QDAYALCRVIKK 503
>AW329169 similar to PIR|E84636|E84 NAM (no apical meristem)-like protein
[imported] - Arabidopsis thaliana, partial (52%)
Length = 631
Score = 181 bits (460), Expect = 2e-46
Identities = 88/168 (52%), Positives = 115/168 (68%), Gaps = 3/168 (1%)
Frame = +3
Query: 2 ESSASSE-LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
ES E LPPGFRFHPTDEELI YL N+ + I ++D+ K +PWELP K++
Sbjct: 9 ESGGKEETLPPGFRFHPTDEELITCYLINKISDSSFTGKAITDIDLNKSEPWELPGKAKM 188
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSG--SKQIGVKKSLVFYKGR 118
E EWYFF+ R+RKYP GVR NRAT +GYWK TG D+ I S+ +G+KK+LVFYKGR
Sbjct: 189 GEKEWYFFNMRDRKYPTGVRTNRATNTGYWKTTGKDREIFDSVTSELVGMKKTLVFYKGR 368
Query: 119 PPKGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGK 166
P+G K++W+MHEYR+ K T + + + D+WV+CRI+KK GK
Sbjct: 369 APRGEKSNWVMHEYRI--HSKSTFR---TNKQDEWVICRIFKKSGSGK 497
>TC86725 similar to GP|9758909|dbj|BAB09485.1 NAM (no apical meristem)-like
protein {Arabidopsis thaliana}, partial (50%)
Length = 1645
Score = 180 bits (456), Expect = 6e-46
Identities = 98/236 (41%), Positives = 139/236 (58%), Gaps = 4/236 (1%)
Frame = +3
Query: 8 ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYF 67
+LPPGFRFHPTD E+IV YL + + A+ I E D+ K +PW+LP K++ E EWYF
Sbjct: 174 DLPPGFRFHPTDAEIIVCYLTEKVKNSKFSATAIGEADLNKCEPWDLPKKAKMGEKEWYF 353
Query: 68 FSPRERKYPNGVRPNRATLSGYWKATGTDKAI----KSGSKQIGVKKSLVFYKGRPPKGV 123
F ++RKYP G+R NRAT SGYWKATG DK I K +G+KK+LVFYKGR PKG
Sbjct: 354 FCQKDRKYPTGMRTNRATESGYWKATGKDKEIYHKGKGIQNLVGMKKTLVFYKGRAPKGE 533
Query: 124 KTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFNDSI 183
KT+W+MHE+RL G K + ++ + D+WV+ R++ K D Q + +
Sbjct: 534 KTNWVMHEFRLEG--KFATHNLPNKEKDEWVVSRVFHK---------NTDVKKPQISSGL 680
Query: 184 ITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDGSTLDFQINNSNIGIDPYVKP 239
+ N G ++++ + SL L+D +Y G T + QI+++ D Y P
Sbjct: 681 LRINSIGHDDLLDYS---SLPPLMDPSYTNDDFK-GITTNQQISSTKSQSDGYYLP 836
>CB894613 similar to GP|16226943|gb AT4g27410/F27G19_10 {Arabidopsis
thaliana}, partial (55%)
Length = 657
Score = 177 bits (448), Expect = 5e-45
Identities = 83/162 (51%), Positives = 101/162 (62%), Gaps = 24/162 (14%)
Frame = +3
Query: 24 VHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFFSPRERKYPNGVRPNR 83
V YLC + II E+D+YKFDPW LP K+ F E EWYFFSPR+RKYPNG RPNR
Sbjct: 3 VQYLCRKVAGHHFSLQIIAEIDLYKFDPWILPSKAIFGEKEWYFFSPRDRKYPNGTRPNR 182
Query: 84 ATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDWIMHEYRLIGSQKQTS- 142
SGYWKATGTDK I + +++G+KK+LVFY G+ PKG KT+WIMHEYRL+ S +
Sbjct: 183 VAGSGYWKATGTDKIITNEGRKVGIKKALVFYVGKAPKGTKTNWIMHEYRLLDSSRNNGG 362
Query: 143 -----------------------KHIGSMRLDDWVLCRIYKK 161
+ ++LDDWVLCRIYKK
Sbjct: 363 TKVLYTFIFFYHAIIIITTLIYLYFVVELQLDDWVLCRIYKK 488
>TC84760 similar to GP|8567777|gb|AAF76349.1| unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 781
Score = 170 bits (431), Expect = 5e-43
Identities = 79/163 (48%), Positives = 111/163 (67%), Gaps = 4/163 (2%)
Frame = +2
Query: 7 SELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFE--ENE 64
+ L PGFRFHPTDEEL+++YL + + KP I EVDIYK +PW+L DKS + + E
Sbjct: 212 TSLAPGFRFHPTDEELVIYYLKRKVSGKPFRFDAIAEVDIYKSEPWDLFDKSRLKTRDQE 391
Query: 65 WYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVK 124
WYFFS ++KY NG R NRATL GYWKATG D+ +K + +G+KK+LVF+ GR P G +
Sbjct: 392 WYFFSALDKKYGNGGRMNRATLQGYWKATGNDRPVKHDLRTVGLKKTLVFHSGRAPDGKR 571
Query: 125 TDWIMHEYRLIGSQKQTSKHIG--SMRLDDWVLCRIYKKKHMG 165
T+W+MHEYRL+ + + + S + +VLC ++ K ++G
Sbjct: 572 TNWVMHEYRLVEEELERERAGAGPSQPQEAFVLCXVFHKNNIG 700
>TC89247 similar to GP|20303588|gb|AAM19015.1 putative NAM (no apical
meristem) protein {Oryza sativa (japonica
cultivar-group)}, partial (39%)
Length = 1255
Score = 167 bits (422), Expect = 5e-42
Identities = 94/235 (40%), Positives = 132/235 (56%), Gaps = 16/235 (6%)
Frame = +2
Query: 11 PGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFFSP 70
PGFRFHPTD+EL+ YL + KP +I E+DIYK+DPW+LP S E E YFF
Sbjct: 251 PGFRFHPTDDELVGFYLRRKVEKKPLRIELIKEIDIYKYDPWDLPKASTVGEKECYFFCL 430
Query: 71 RERKYPNGVRPNRATLSGYWKATGTDK---AIKSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
R RKY N +RPNR T SG+WKATG DK +K + IG+KKSLV+Y+G KG KTDW
Sbjct: 431 RGRKYRNSIRPNRVTGSGFWKATGIDKPIYCVKESHECIGLKKSLVYYRGSAGKGTKTDW 610
Query: 128 IMHEYRL----IGSQKQTSKHIGSMR-LDDWVLCRIYKK--KHMGKTLQQKEDYSTHQF- 179
+MHE+RL S Q H ++ + W LCRI+K+ + T K+D F
Sbjct: 611 MMHEFRLPPKGKTSNNQVLNHTNDVQEAEVWTLCRIFKRIPSYKKYTPNVKKDSEAAAFT 790
Query: 180 -----NDSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDGSTLDFQINNS 229
+ + ++D+ + + + T S +++ N+ +G +D + NNS
Sbjct: 791 INSSSSKTCSLDSDNTKPQYLTFTNS---QHVVQQNHETKPFINGHVVDQRFNNS 946
>TC77157 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like protein
{Arabidopsis thaliana}, partial (66%)
Length = 1998
Score = 167 bits (422), Expect = 5e-42
Identities = 74/123 (60%), Positives = 91/123 (73%)
Frame = +3
Query: 21 ELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFFSPRERKYPNGVR 80
EL++HYLC + TS+P II E+D+YK+DPW+LP + + E EWYFFSPR+RKYPNG R
Sbjct: 3 ELVMHYLCRKCTSQPIAVPIIAEIDLYKYDPWDLPGMATYGEKEWYFFSPRDRKYPNGSR 182
Query: 81 PNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDWIMHEYRLIGSQKQ 140
PNRA SGYWKATG DK I K +G+KK+LVFY G+ PKG KT+WIMHEYRL +
Sbjct: 183 PNRAAGSGYWKATGADKPI-GHPKPVGIKKALVFYAGKAPKGDKTNWIMHEYRLADVDRS 359
Query: 141 TSK 143
K
Sbjct: 360 IRK 368
Score = 32.0 bits (71), Expect = 0.26
Identities = 12/14 (85%), Positives = 13/14 (92%)
Frame = +3
Query: 149 RLDDWVLCRIYKKK 162
+LDDWVLCRIY KK
Sbjct: 849 QLDDWVLCRIYNKK 890
>TC82268 homologue to GP|20197337|gb|AAC78526.2 NAM (no apical
meristem)-like protein {Arabidopsis thaliana}, partial
(40%)
Length = 862
Score = 165 bits (417), Expect = 2e-41
Identities = 77/152 (50%), Positives = 106/152 (69%), Gaps = 1/152 (0%)
Frame = +2
Query: 11 PGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFFSP 70
PGFRFHPT+EEL+ YL + K +I +D+Y++DPWELP + E EWYF+ P
Sbjct: 260 PGFRFHPTEEELVEFYLRRKVEGKRFNVELITFLDLYRYDPWELPALAAIGEKEWYFYVP 439
Query: 71 RERKYPNGVRPNRATLSGYWKATGTDKAIKSGS-KQIGVKKSLVFYKGRPPKGVKTDWIM 129
R+RKY NG RPNR T SGYWKATG D+ I++ + + IG+KK+LVFY G+ PKG++T WIM
Sbjct: 440 RDRKYRNGDRPNRVTTSGYWKATGADRMIRTENFRSIGLKKTLVFYSGKAPKGIRTSWIM 619
Query: 130 HEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKK 161
+EYRL Q +T ++ + + LCR+YK+
Sbjct: 620 NEYRL--PQHETERY----QKGEISLCRVYKR 697
>BG646875 similar to GP|21593065|gb NAM / CUC2-like protein {Arabidopsis
thaliana}, partial (42%)
Length = 760
Score = 164 bits (414), Expect = 4e-41
Identities = 80/177 (45%), Positives = 114/177 (64%), Gaps = 7/177 (3%)
Frame = +3
Query: 7 SELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWY 66
+ LPPGFRF+P+DEEL++HYL + T++ + E+D++ +PW+LP+ ++ EWY
Sbjct: 141 ASLPPGFRFYPSDEELVLHYLYKKITNEDVLKGTLEEIDLHTCEPWQLPEVAKLNATEWY 320
Query: 67 FFSPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKKSLVFYKGRPPKGVK 124
FFS R+RKY G R NRAT SGYWKATG D+ + + +G++K+LVFY+ R P G+K
Sbjct: 321 FFSFRDRKYATGFRTNRATTSGYWKATGKDRTVFDPITHEVVGMRKTLVFYRNRAPNGIK 500
Query: 125 TDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKT-----LQQKEDYST 176
T WIMHE+RL + H+ +DWVLCR++ H GKT L +E Y T
Sbjct: 501 TGWIMHEFRL------ETPHMPPK--EDWVLCRVF---HKGKTENNAKLSPQEMYET 638
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,579,547
Number of Sequences: 36976
Number of extensions: 142000
Number of successful extensions: 746
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 680
length of query: 271
length of database: 9,014,727
effective HSP length: 95
effective length of query: 176
effective length of database: 5,502,007
effective search space: 968353232
effective search space used: 968353232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC140030.7


