

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
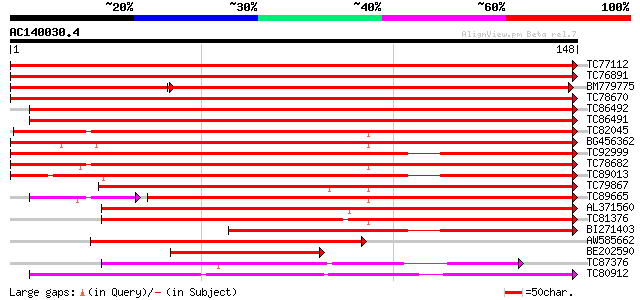
Query= AC140030.4 - phase: 0 /pseudo
(148 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77112 homologue to GP|21615409|emb|CAD33924. alpha-expansin 4 ... 322 5e-89
TC76891 homologue to GP|21615407|emb|CAD33923. alpha-expansin 3 ... 285 4e-78
BM779775 homologue to GP|21615407|em alpha-expansin 3 {Cicer ari... 216 2e-73
TC78670 homologue to GP|11932092|emb|CAC19184. expansin {Cicer a... 251 1e-67
TC86492 homologue to GP|11907554|dbj|BAB19676. expansin {Prunus ... 250 2e-67
TC86491 homologue to GP|11907554|dbj|BAB19676. expansin {Prunus ... 250 2e-67
TC82045 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus ce... 230 2e-61
BG456362 homologue to GP|732905|emb| homology with pollen allerg... 228 9e-61
TC92999 similar to GP|11932089|emb|CAC19183. expansin {Cicer ari... 227 1e-60
TC78682 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus ce... 223 2e-59
TC89013 homologue to GP|11932089|emb|CAC19183. expansin {Cicer a... 223 2e-59
TC79867 similar to PIR|T10083|T10083 expansin S2 precursor - cuc... 209 3e-55
TC89665 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus ce... 193 4e-53
AL371560 similar to SP|Q9LN94|EXP7 Alpha-expansin 7 precursor (A... 167 1e-42
TC81376 similar to SP|Q9LN94|EXP7_ARATH Alpha-expansin 7 precurs... 163 3e-41
BI271403 similar to SP|Q38864|EXP5 Alpha-expansin 5 precursor (A... 147 2e-36
AW585662 similar to GP|16923359|gb| alpha-expansin 6 precursor {... 130 3e-31
BE202590 similar to GP|18057102|gb| putative alpha-expansin prot... 74 3e-14
TC87376 similar to GP|21592834|gb|AAM64784.1 putative beta-expan... 66 5e-12
TC80912 similar to SP|O23547|EXR1_ARATH Expansin-related protein... 64 2e-11
>TC77112 homologue to GP|21615409|emb|CAD33924. alpha-expansin 4 {Cicer
arietinum}, complete
Length = 859
Score = 322 bits (824), Expect = 5e-89
Identities = 148/148 (100%), Positives = 148/148 (100%)
Frame = +1
Query: 1 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 60
MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT
Sbjct: 37 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 216
Query: 61 AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL 120
AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL
Sbjct: 217 AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL 396
Query: 121 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
QHFDLAQPVFLRIAQYKAGIVPVDFRRV
Sbjct: 397 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 480
>TC76891 homologue to GP|21615407|emb|CAD33923. alpha-expansin 3 {Cicer
arietinum}, complete
Length = 1432
Score = 285 bits (730), Expect = 4e-78
Identities = 129/148 (87%), Positives = 137/148 (92%)
Frame = +3
Query: 1 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 60
MA IGL ++ LTMFSS Y YGGGW NAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT
Sbjct: 123 MALIGLFIMVFLTMFSSAYGYGGGWVNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 302
Query: 61 AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL 120
AALSTALFN+GLSCGSC+EIRCA D +WCLPGSI+VTATNFCPPN ALPNN+GGWCNPPL
Sbjct: 303 AALSTALFNSGLSCGSCFEIRCAGDRKWCLPGSILVTATNFCPPNTALPNNNGGWCNPPL 482
Query: 121 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
QHFDLAQPVFLRIAQYKAGIVPV +RRV
Sbjct: 483 QHFDLAQPVFLRIAQYKAGIVPVSYRRV 566
>BM779775 homologue to GP|21615407|em alpha-expansin 3 {Cicer arietinum},
partial (59%)
Length = 761
Score = 216 bits (549), Expect(2) = 2e-73
Identities = 95/106 (89%), Positives = 102/106 (95%)
Frame = +1
Query: 42 MGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNF 101
+GGACGYGNLYSQGYGTNTAALSTALFN+GLSCGSC+EIRCA D +WCLPGSI+VTATNF
Sbjct: 289 LGGACGYGNLYSQGYGTNTAALSTALFNSGLSCGSCFEIRCAGDRKWCLPGSILVTATNF 468
Query: 102 CPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRR 147
CPPN ALPNN+GGWCNPPLQHFDLAQPVFLRIAQYKAGIVPV +RR
Sbjct: 469 CPPNTALPNNNGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVSYRR 606
Score = 75.9 bits (185), Expect(2) = 2e-73
Identities = 34/43 (79%), Positives = 36/43 (83%)
Frame = +2
Query: 1 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMG 43
MA IGL ++ LTMFSS Y YGGGW NAHATFYGGSDASGTMG
Sbjct: 59 MALIGLFIMVFLTMFSSAYGYGGGWVNAHATFYGGSDASGTMG 187
>TC78670 homologue to GP|11932092|emb|CAC19184. expansin {Cicer arietinum},
complete
Length = 1043
Score = 251 bits (640), Expect = 1e-67
Identities = 112/148 (75%), Positives = 128/148 (85%)
Frame = +1
Query: 1 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 60
+A + LV++C V A GGW +AHATFYGG DASGTMGGACGYGNLYSQGYGTNT
Sbjct: 85 IALVILVVLCINMNLQGVIADYGGWESAHATFYGGGDASGTMGGACGYGNLYSQGYGTNT 264
Query: 61 AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL 120
AALSTALFNNGLSCGSCYE+RC +D RWC PGSI+VTATNFCPPN +LPNN+GGWCNPPL
Sbjct: 265 AALSTALFNNGLSCGSCYEMRCNDDPRWCKPGSIIVTATNFCPPNPSLPNNNGGWCNPPL 444
Query: 121 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
QHFD+A+P +L+IA+Y+AGIVPV FRRV
Sbjct: 445 QHFDMAEPAYLQIAEYRAGIVPVSFRRV 528
>TC86492 homologue to GP|11907554|dbj|BAB19676. expansin {Prunus persica},
partial (95%)
Length = 1257
Score = 250 bits (638), Expect = 2e-67
Identities = 113/143 (79%), Positives = 124/143 (86%)
Frame = +3
Query: 6 LVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
L V SL + + YGGGW NAHATFYGG DASGTMGGACGYGNLYSQGYGTNTAALST
Sbjct: 213 LFFVLSLCLKGTFGDYGGGWENAHATFYGGGDASGTMGGACGYGNLYSQGYGTNTAALST 392
Query: 66 ALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
ALFNNGLSCGSCYE++C +D +WCLPGSI+VTATNFCPPN A N +GGWCNPPLQHFDL
Sbjct: 393 ALFNNGLSCGSCYEMKCNSDPKWCLPGSILVTATNFCPPNFAESNTNGGWCNPPLQHFDL 572
Query: 126 AQPVFLRIAQYKAGIVPVDFRRV 148
A+P FL+IAQYKAGIVP+ FRRV
Sbjct: 573 AEPAFLQIAQYKAGIVPISFRRV 641
>TC86491 homologue to GP|11907554|dbj|BAB19676. expansin {Prunus persica},
partial (95%)
Length = 1166
Score = 250 bits (638), Expect = 2e-67
Identities = 113/143 (79%), Positives = 124/143 (86%)
Frame = +2
Query: 6 LVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
L V SL + + YGGGW NAHATFYGG DASGTMGGACGYGNLYSQGYGTNTAALST
Sbjct: 104 LFFVLSLCLRGTFGDYGGGWENAHATFYGGGDASGTMGGACGYGNLYSQGYGTNTAALST 283
Query: 66 ALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
ALFNNGLSCGSCYE++C +D +WCLPGSI+VTATNFCPPN A N +GGWCNPPLQHFDL
Sbjct: 284 ALFNNGLSCGSCYEMKCNSDPKWCLPGSILVTATNFCPPNFAESNTNGGWCNPPLQHFDL 463
Query: 126 AQPVFLRIAQYKAGIVPVDFRRV 148
A+P FL+IAQYKAGIVP+ FRRV
Sbjct: 464 AEPAFLQIAQYKAGIVPISFRRV 532
>TC82045 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus cerasus},
partial (91%)
Length = 814
Score = 230 bits (586), Expect = 2e-61
Identities = 106/149 (71%), Positives = 123/149 (82%), Gaps = 2/149 (1%)
Frame = +3
Query: 2 AFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTA 61
+F+ L+L + VY+ GG W AHATFYGGSDASGTMGGACGYGNLYSQGYG NTA
Sbjct: 42 SFLALLLTSTEARLPGVYS-GGAWQTAHATFYGGSDASGTMGGACGYGNLYSQGYGVNTA 218
Query: 62 ALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNNDGGWCNPP 119
ALSTALFNNGLSCGSC+E++C+ND WC PG SI+VTATNFCPPN A +++GGWCNPP
Sbjct: 219 ALSTALFNNGLSCGSCFELKCSNDPSWCHPGSPSILVTATNFCPPNFAQASDNGGWCNPP 398
Query: 120 LQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
HFDLA P+FL+IAQY+AGIVPV +RRV
Sbjct: 399 RPHFDLAMPMFLKIAQYRAGIVPVSYRRV 485
>BG456362 homologue to GP|732905|emb| homology with pollen allergens {Pisum
sativum}, partial (79%)
Length = 655
Score = 228 bits (580), Expect = 9e-61
Identities = 109/157 (69%), Positives = 129/157 (81%), Gaps = 9/157 (5%)
Frame = +2
Query: 1 MAFIGLVLVCSL-TMFSSVYAY------GGGWTNAHATFYGGSDASGTMGGACGYGNLYS 53
MA I L+ + SL ++F++ A GG WT+AHATFYGGSDASGTMGGACGYGNLYS
Sbjct: 29 MAGILLLTIASLASLFAATTARIPGVYSGGPWTSAHATFYGGSDASGTMGGACGYGNLYS 208
Query: 54 QGYGTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNN 111
QGYG NTAALSTALFNNGLSCG+C+E++C D RWC PG SI++TATNFCPPN A P++
Sbjct: 209 QGYGVNTAALSTALFNNGLSCGACFELKCDQDPRWCNPGNPSILITATNFCPPNFAQPSD 388
Query: 112 DGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
+GGWCNPP HFDLA P+FL+IAQY+AGIVPV +RRV
Sbjct: 389 NGGWCNPPRPHFDLAMPMFLKIAQYRAGIVPVAYRRV 499
>TC92999 similar to GP|11932089|emb|CAC19183. expansin {Cicer arietinum},
partial (96%)
Length = 767
Score = 227 bits (579), Expect = 1e-60
Identities = 107/148 (72%), Positives = 119/148 (80%)
Frame = +2
Query: 1 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 60
+ F+ L ++ SL + Y GW NAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT
Sbjct: 23 LGFLVLGIIFSLISYVHGYGRNWGWINAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 202
Query: 61 AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL 120
AALSTALFNNGLSCG+CYEI+CA+D +WCL GSIVVTATNFCPP GGWC+PP
Sbjct: 203 AALSTALFNNGLSCGACYEIKCASDPKWCLHGSIVVTATNFCPP--------GGWCDPPN 358
Query: 121 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
HFDL+QPVF IAQYKAGIVPV +RRV
Sbjct: 359 HHFDLSQPVFQHIAQYKAGIVPVVYRRV 442
>TC78682 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus cerasus},
partial (91%)
Length = 1236
Score = 223 bits (568), Expect = 2e-59
Identities = 104/154 (67%), Positives = 125/154 (80%), Gaps = 6/154 (3%)
Frame = +1
Query: 1 MAFIGLVLVCSLTMFSS----VYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGY 56
M +I +++ S+ + + VY+ GG W +AHATFYGGSDASGTMGGACGYGNLYSQGY
Sbjct: 109 MLYIAFIIITSMYIVQARIPGVYS-GGQWQDAHATFYGGSDASGTMGGACGYGNLYSQGY 285
Query: 57 GTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNNDGG 114
G NTAALSTALFNNGLSCG+C+EI+CAND WC G SI +TATNFCPPN A +++GG
Sbjct: 286 GVNTAALSTALFNNGLSCGACFEIKCANDKEWCHSGSPSIFITATNFCPPNFAQASDNGG 465
Query: 115 WCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
WCNPP HFDLA P+FL+IA+Y+AGIVPV +RRV
Sbjct: 466 WCNPPRPHFDLAMPMFLKIAEYRAGIVPVAYRRV 567
>TC89013 homologue to GP|11932089|emb|CAC19183. expansin {Cicer arietinum},
partial (94%)
Length = 1318
Score = 223 bits (568), Expect = 2e-59
Identities = 107/150 (71%), Positives = 119/150 (79%), Gaps = 2/150 (1%)
Frame = +3
Query: 1 MAFIGLVLVCSLTMFSSVYAYGG--GWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGT 58
M+ +GL+L M S V GG W NAHATFYGGSDASGTMGGACGYGNLYSQGYGT
Sbjct: 6 MSLLGLLLAI-FYMVSHVNGRGGHASWINAHATFYGGSDASGTMGGACGYGNLYSQGYGT 182
Query: 59 NTAALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNP 118
NTAALSTALFNNGLSCG+CYEI+C ND +WC PGSI+VTATNFCPP GGWC+P
Sbjct: 183 NTAALSTALFNNGLSCGACYEIKCVNDPQWCNPGSIIVTATNFCPP--------GGWCDP 338
Query: 119 PLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
P HFDL+QP+F IAQY+AGIVPV +RRV
Sbjct: 339 PNHHFDLSQPIFQHIAQYRAGIVPVAYRRV 428
>TC79867 similar to PIR|T10083|T10083 expansin S2 precursor - cucumber,
partial (87%)
Length = 1211
Score = 209 bits (533), Expect = 3e-55
Identities = 96/128 (75%), Positives = 107/128 (83%), Gaps = 3/128 (2%)
Frame = +2
Query: 24 GWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRC- 82
GWTNAHATFYGGSDA GTMGGACGYGNLYSQGYGT TAALSTALFN+G SCG CY+I C
Sbjct: 14 GWTNAHATFYGGSDAEGTMGGACGYGNLYSQGYGTRTAALSTALFNDGASCGQCYKIICD 193
Query: 83 -ANDHRWCLPG-SIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGI 140
D RWC+ G SI +TATNFCPPN LPN+DGGWCNPPL+HFD+AQP + +I Y+ GI
Sbjct: 194 YKTDPRWCIKGRSITITATNFCPPNFDLPNDDGGWCNPPLKHFDMAQPAWEKIGIYRGGI 373
Query: 141 VPVDFRRV 148
VPV F+RV
Sbjct: 374 VPVLFQRV 397
>TC89665 similar to GP|13898655|gb|AAK48848.1 expansin {Prunus cerasus},
partial (92%)
Length = 1006
Score = 193 bits (491), Expect(2) = 4e-53
Identities = 88/114 (77%), Positives = 100/114 (87%), Gaps = 2/114 (1%)
Frame = +3
Query: 37 DASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SI 94
DASGTMGGACGYGNLYSQGYG NTAALSTALFNNGLSCGSC+E++ AND +WC G SI
Sbjct: 204 DASGTMGGACGYGNLYSQGYGVNTAALSTALFNNGLSCGSCFELKWANDKQWCHSGSPSI 383
Query: 95 VVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
+TATNFCPPN A P+++GGWCNPP HFDLA P+FL+IA+Y+AGIVPV FRRV
Sbjct: 384 FITATNFCPPNFAQPSDNGGWCNPPRPHFDLAMPMFLKIAEYRAGIVPVAFRRV 545
Score = 30.4 bits (67), Expect(2) = 4e-53
Identities = 17/35 (48%), Positives = 20/35 (56%), Gaps = 6/35 (17%)
Frame = +1
Query: 6 LVLVCSLTMFS------SVYAYGGGWTNAHATFYG 34
+ L+ SLTM+ VY GG W AHATFYG
Sbjct: 94 VTLIMSLTMWMVDARIPGVYN-GGAWQTAHATFYG 195
>AL371560 similar to SP|Q9LN94|EXP7 Alpha-expansin 7 precursor (At-EXP7)
(AtEx7) (Ath-ExpAlpha-1.26). [Mouse-ear cress], partial
(54%)
Length = 463
Score = 167 bits (424), Expect = 1e-42
Identities = 75/125 (60%), Positives = 95/125 (76%), Gaps = 1/125 (0%)
Frame = +2
Query: 25 WTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCAN 84
W+ AHATFYG AS TMGGACGYGNL+ GYGT+T ALS+ LFNNG +CG+C++I+C
Sbjct: 8 WSLAHATFYGDETASETMGGACGYGNLFQNGYGTDTVALSSTLFNNGYACGTCFQIKCYQ 187
Query: 85 DHR-WCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPV 143
+ VTATN CPPN + P+++GGWCNPP HFD+A+P F++IAQ+KAGIVPV
Sbjct: 188 SSACYRNVAFTTVTATNLCPPNWSKPSDNGGWCNPPRVHFDMAKPAFMKIAQWKAGIVPV 367
Query: 144 DFRRV 148
+RRV
Sbjct: 368 MYRRV 382
>TC81376 similar to SP|Q9LN94|EXP7_ARATH Alpha-expansin 7 precursor
(At-EXP7) (AtEx7) (Ath-ExpAlpha-1.26). [Mouse-ear
cress], partial (80%)
Length = 997
Score = 163 bits (412), Expect = 3e-41
Identities = 75/126 (59%), Positives = 92/126 (72%), Gaps = 2/126 (1%)
Frame = +1
Query: 25 WTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCAN 84
WT AHATFYG AS TMGGACGYGNL+ GYGT+TAALS+ LFNNG +CG+CY+I+C
Sbjct: 118 WTLAHATFYGDETASATMGGACGYGNLFVNGYGTDTAALSSTLFNNGYACGTCYQIKCVQ 297
Query: 85 DHRWCLPG--SIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVP 142
C VTATN CPPN + +++GGWCNPP HFD+ +P F++IAQ+KAGI P
Sbjct: 298 SSA-CNTNVPYTTVTATNICPPNWSQASDNGGWCNPPRSHFDMPKPAFMKIAQWKAGIRP 474
Query: 143 VDFRRV 148
V +R V
Sbjct: 475 VMYRTV 492
>BI271403 similar to SP|Q38864|EXP5 Alpha-expansin 5 precursor (At-EXP5)
(AtEx5) (Ath-ExpAlpha-1.4). [Mouse-ear cress], partial
(65%)
Length = 664
Score = 147 bits (371), Expect = 2e-36
Identities = 69/91 (75%), Positives = 76/91 (82%)
Frame = +2
Query: 58 TNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCN 117
TNTAALSTALFNNGLSCG+CYEI+CA+D +WCL GSIVVTATNFCPP GGWC+
Sbjct: 143 TNTAALSTALFNNGLSCGACYEIKCASDPKWCLHGSIVVTATNFCPP--------GGWCD 298
Query: 118 PPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
PP HFDL+QPVF IAQYKAGIVPV +RRV
Sbjct: 299 PPNHHFDLSQPVFQHIAQYKAGIVPVVYRRV 391
>AW585662 similar to GP|16923359|gb| alpha-expansin 6 precursor {Cucumis
sativus}, partial (33%)
Length = 355
Score = 130 bits (326), Expect = 3e-31
Identities = 57/72 (79%), Positives = 61/72 (84%)
Frame = +1
Query: 22 GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIR 81
GG W AHATFYGGSDASG MGGACGYGN SQGYG NTAALSTALFN GLSCGSC+E++
Sbjct: 136 GGAWQTAHATFYGGSDASGAMGGACGYGN*NSQGYGVNTAALSTALFNTGLSCGSCFELK 315
Query: 82 CANDHRWCLPGS 93
CAND +WC GS
Sbjct: 316 CANDKQWCHSGS 351
>BE202590 similar to GP|18057102|gb| putative alpha-expansin protein {Oryza
sativa (japonica cultivar-group)}, partial (20%)
Length = 596
Score = 73.6 bits (179), Expect = 3e-14
Identities = 29/40 (72%), Positives = 36/40 (89%)
Frame = +2
Query: 43 GGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRC 82
GGACGYGNL+ GYGT+TAALS+ LFNNG +CG+CY+I+C
Sbjct: 476 GGACGYGNLFVNGYGTDTAALSSTLFNNGYACGTCYQIKC 595
Score = 33.5 bits (75), Expect = 0.032
Identities = 14/19 (73%), Positives = 14/19 (73%)
Frame = +3
Query: 25 WTNAHATFYGGSDASGTMG 43
WT AHATFYG AS TMG
Sbjct: 105 WTLAHATFYGDETASATMG 161
>TC87376 similar to GP|21592834|gb|AAM64784.1 putative
beta-expansin/allergen protein {Arabidopsis thaliana},
partial (81%)
Length = 976
Score = 66.2 bits (160), Expect = 5e-12
Identities = 36/111 (32%), Positives = 52/111 (46%), Gaps = 1/111 (0%)
Frame = +2
Query: 25 WTNAHATFYGGSDASGTMGGACGYGNLYS-QGYGTNTAALSTALFNNGLSCGSCYEIRCA 83
W AT+YG ++ G+ GGACGYG+L + A+ L+ G CG CY+++C
Sbjct: 200 WHPGTATWYGEAEGDGSRGGACGYGSLVDVKPLRARVGAVGPVLYKKGEGCGECYKVKCL 379
Query: 84 NDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIA 134
DH C ++ V T+ CP C HFDL+ F +A
Sbjct: 380 -DHTICSKRAVTVIITDECP-----------GCPSDRTHFDLSGAAFGHMA 496
>TC80912 similar to SP|O23547|EXR1_ARATH Expansin-related protein 1
precursor (At-EXPR1) (Ath-ExpBeta-3.1). [Mouse-ear
cress], partial (35%)
Length = 764
Score = 64.3 bits (155), Expect = 2e-11
Identities = 38/143 (26%), Positives = 66/143 (45%)
Frame = +2
Query: 6 LVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
L LVC + + + +T++ AT+YG D GT GACG+G Y + A +
Sbjct: 107 LGLVCIILLLPQLCTSQDSFTDSRATYYGSPDCYGTPRGACGFGE-YGRTVNDGNVAGVS 283
Query: 66 ALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
L+ NG CG+CY++RC ++C V T++ + + P + L
Sbjct: 284 KLWKNGTGCGACYQVRC-KIPQYCDENGATVVVTDYGEGDRT------DFIMSPRGYAKL 442
Query: 126 AQPVFLRIAQYKAGIVPVDFRRV 148
+ +K G+V ++++RV
Sbjct: 443 GRNADASAELFKYGVVDIEYKRV 511
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.140 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,993,138
Number of Sequences: 36976
Number of extensions: 101911
Number of successful extensions: 652
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 602
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 629
length of query: 148
length of database: 9,014,727
effective HSP length: 87
effective length of query: 61
effective length of database: 5,797,815
effective search space: 353666715
effective search space used: 353666715
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC140030.4


