

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140025.4 + phase: 0
(277 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
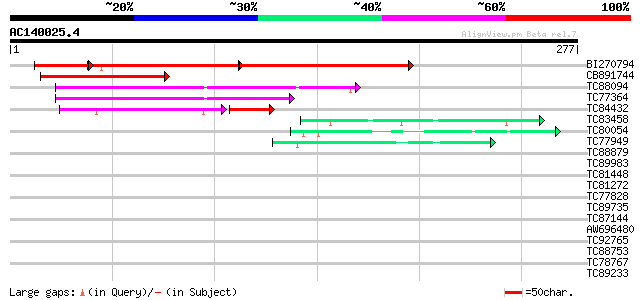
Score E
Sequences producing significant alignments: (bits) Value
BI270794 similar to GP|18921326|gb putative polypyrimidine tract... 182 3e-96
CB891744 similar to GP|18921326|gb| putative polypyrimidine trac... 121 3e-28
TC88094 similar to GP|20160852|dbj|BAB89791. contains EST AU0557... 85 3e-17
TC77364 similar to GP|20160852|dbj|BAB89791. contains EST AU0557... 84 6e-17
TC84432 similar to GP|9757999|dbj|BAB08421.1 polypyrimidine trac... 64 9e-14
TC83458 homologue to PIR|T07796|T07796 DNA-directed RNA polymera... 44 5e-05
TC80054 SP|Q28009|FUS_BOVIN RNA-binding protein FUS (Pigpen prot... 44 9e-05
TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {A... 41 6e-04
TC88879 weakly similar to PIR|T04011|T04011 hypothetical protein... 40 0.001
TC89983 similar to GP|14331087|emb|CAC40974. hypothetical protei... 39 0.002
TC81448 weakly similar to OMNI|NT01MC3624 toxin secretion ATP-bi... 38 0.004
TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 ... 37 0.011
TC77828 similar to GP|16648867|gb|AAL24285.1 HSP associated prot... 37 0.011
TC89735 similar to GP|19569060|gb|AAL92018.1 UPF1 {Arabidopsis t... 36 0.014
TC87144 similar to GP|21595622|gb|AAM66118.1 unknown {Arabidopsi... 35 0.024
AW696480 weakly similar to SP|P03211|EBN1_ EBNA-1 nuclear protei... 35 0.041
TC92765 similar to EGAD|119550|127787 hypothetical protein F49E1... 34 0.070
TC88753 similar to PIR|T10586|T10586 small nuclear ribonucleopro... 34 0.070
TC78767 similar to PIR|S59117|S59117 small nuclear ribonucleopro... 33 0.12
TC89233 homologue to GP|18734|emb|CAA36734.1|| DNA-directed RNA ... 33 0.16
>BI270794 similar to GP|18921326|gb putative polypyrimidine tract-binding
protein {Oryza sativa}, partial (26%)
Length = 659
Score = 182 bits (463), Expect(3) = 3e-96
Identities = 83/85 (97%), Positives = 84/85 (98%)
Frame = +2
Query: 113 VKAFSDKSRDYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSW 172
+ AFSDKSRDYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSW
Sbjct: 404 LNAFSDKSRDYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSW 583
Query: 173 DLTQHAVRPGYVPVPGAYPGQTGAF 197
DLTQHAVRPGYVPVPGAYPGQTGAF
Sbjct: 584 DLTQHAVRPGYVPVPGAYPGQTGAF 658
Score = 148 bits (373), Expect(3) = 3e-96
Identities = 74/79 (93%), Positives = 75/79 (94%), Gaps = 3/79 (3%)
Frame = +3
Query: 39 QYAVT---VDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCIYD 95
+YAV VDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCIYD
Sbjct: 72 EYAVCLLPVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCIYD 251
Query: 96 GGYCKLHLSYSRHTDLNVK 114
GGYCKLHLSYSRHTDLNVK
Sbjct: 252 GGYCKLHLSYSRHTDLNVK 308
Score = 60.1 bits (144), Expect(3) = 3e-96
Identities = 29/29 (100%), Positives = 29/29 (100%)
Frame = +1
Query: 13 AQPAIGPDGKRIETESNVLLASIENMQYA 41
AQPAIGPDGKRIETESNVLLASIENMQYA
Sbjct: 1 AQPAIGPDGKRIETESNVLLASIENMQYA 87
>CB891744 similar to GP|18921326|gb| putative polypyrimidine tract-binding
protein {Oryza sativa}, partial (13%)
Length = 366
Score = 121 bits (304), Expect = 3e-28
Identities = 61/63 (96%), Positives = 62/63 (97%)
Frame = +1
Query: 16 AIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQY 75
AIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQY
Sbjct: 1 AIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQY 180
Query: 76 PDV 78
P +
Sbjct: 181PGI 189
>TC88094 similar to GP|20160852|dbj|BAB89791. contains EST
AU055715(S20012)~similar to nuclear
ribonucleoprotein~unknown protein, partial (22%)
Length = 890
Score = 85.1 bits (209), Expect = 3e-17
Identities = 56/158 (35%), Positives = 81/158 (50%), Gaps = 9/158 (5%)
Frame = +1
Query: 23 RIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAA 82
R + + +LL +I ++ Y +TVDV+ VFS G+V+KI F+K+ QALIQY ++
Sbjct: 358 RGDEPNRILLVTIHHVLYPITVDVLYQVFSPHGSVEKIVTFQKSAGFQALIQYQSQQSSI 537
Query: 83 AAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQNPQAAPMY 142
AR AL+G IYD G C+L + +S +L V +D+SRD+T P +P P Y
Sbjct: 538 TARTALQGRNIYD-GCCQLDIQFSNLDELQVNYNNDRSRDFTNPNLPTEQKGRPPQLG-Y 711
Query: 143 PTNSPAYQTQVPGGSPAYQTQVP---------GGQVPS 171
Y Q G Q+P GG +PS
Sbjct: 712 GDAGNMYGVQGSGPRTVGYPQMPNAAAIAAAFGGSLPS 825
>TC77364 similar to GP|20160852|dbj|BAB89791. contains EST
AU055715(S20012)~similar to nuclear
ribonucleoprotein~unknown protein, partial (50%)
Length = 2480
Score = 84.0 bits (206), Expect = 6e-17
Identities = 46/117 (39%), Positives = 68/117 (57%)
Frame = +2
Query: 23 RIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAA 82
R + + +LL ++ + Y +TVDV+ VFS G V+KI F+K+ QALIQY +A
Sbjct: 1004 REDEPNRILLVTVHQVLYPMTVDVLQQVFSPHGFVEKIVTFQKSAGFQALIQYETRQSAV 1183
Query: 83 AAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQNPQAA 139
AR AL+G +YD G C+L + +S +L V +D+SRD+T P +P P A
Sbjct: 1184 TARGALQGRNVYD-GCCQLDIQFSNLDELQVNYNNDRSRDFTNPNLPTDQKGRPSQA 1351
Score = 28.1 bits (61), Expect = 3.9
Identities = 16/54 (29%), Positives = 27/54 (49%)
Frame = +2
Query: 35 IENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREAL 88
+ ++ + D I ++ G + +FE NG+ QAL+Q+ T A EAL
Sbjct: 1838 MSSLPQEIIEDEIASLLQEHGIIVNCKVFEMNGKKQALVQF---ETEEEATEAL 1990
>TC84432 similar to GP|9757999|dbj|BAB08421.1 polypyrimidine tract-binding
RNA transport protein-like {Arabidopsis thaliana},
partial (50%)
Length = 685
Score = 63.5 bits (153), Expect(2) = 9e-14
Identities = 37/89 (41%), Positives = 52/89 (57%), Gaps = 7/89 (7%)
Frame = +3
Query: 25 ETESNVLLASIENMQYA-VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
+ NVLL ++E V++DV++ VFSAFG V KI FEK QAL+Q+ D TA +
Sbjct: 345 DVAGNVLLVTVEGEDARLVSIDVLHLVFSAFGFVHKITTFEKTAGFQALVQFSDAETATS 524
Query: 84 AREALEGHCI------YDGGYCKLHLSYS 106
A++AL+G I G C L ++YS
Sbjct: 525 AKDALDGRSIPRYLLSEHVGPCTLKITYS 611
Score = 30.0 bits (66), Expect(2) = 9e-14
Identities = 13/22 (59%), Positives = 16/22 (72%)
Frame = +1
Query: 108 HTDLNVKAFSDKSRDYTVPLVP 129
H+DL VK S +SRDYT P +P
Sbjct: 616 HSDLTVKFQSHRSRDYTNPYLP 681
>TC83458 homologue to PIR|T07796|T07796 DNA-directed RNA polymerase (EC
2.7.7.6) largest chain - soybean (fragment), partial
(63%)
Length = 1220
Score = 44.3 bits (103), Expect = 5e-05
Identities = 39/125 (31%), Positives = 46/125 (36%), Gaps = 6/125 (4%)
Frame = +2
Query: 143 PTNSPAYQTQVPG---GSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAY-PGQTGAFP 198
PT+SP Y PG SP Y PG S + PGY P Y P G P
Sbjct: 812 PTSSPGYSPSSPGYSPSSPGYSPTSPGYSPTSPGYS--PTSPGYSPTSPTYSPSSPGYSP 985
Query: 199 TMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGS--SPHMQQNLGAQGMVR 256
T P+Y + + SP + S P +P PS TS S SP A
Sbjct: 986 TSPAYSPTS--PSYSPTSPSYSPTSPSYSPTSPSYSPTSSSYSPTSPAYSPTSSAYSPTS 1159
Query: 257 PGAPP 261
P P
Sbjct: 1160PAYSP 1174
Score = 37.4 bits (85), Expect = 0.006
Identities = 31/92 (33%), Positives = 40/92 (42%), Gaps = 6/92 (6%)
Frame = +2
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAY-PGQTGAF 197
+P Y +SP Y P SP + P PS+ T P Y P +Y P +
Sbjct: 947 SPTYSPSSPGYSPTSPAYSPTSPSYSPTS--PSYSPTS----PSYSPTSPSYSPTSSSYS 1108
Query: 198 PTMPSY---GSAAMPT--ASSPLAQSSHPGAP 224
PT P+Y SA PT A SP + S P +P
Sbjct: 1109 PTSPAYSPTSSAYSPTSPAYSPTSPSYSPTSP 1204
Score = 31.6 bits (70), Expect = 0.35
Identities = 27/92 (29%), Positives = 35/92 (37%), Gaps = 4/92 (4%)
Frame = +2
Query: 116 FSDKSRDYTVPLVPAPVWQNPQ---AAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSW 172
+S S Y+ P PA +P +P Y SP+Y P SP + P
Sbjct: 956 YSPSSPGYS-PTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSSSYSP------- 1111
Query: 173 DLTQHAVRPGYVPVPGAYPGQTGAF-PTMPSY 203
P Y P AY + A+ PT PSY
Sbjct: 1112 ------TSPAYSPTSSAYSPTSPAYSPTSPSY 1189
>TC80054 SP|Q28009|FUS_BOVIN RNA-binding protein FUS (Pigpen protein).
[Bovine] {Bos taurus}, partial (4%)
Length = 988
Score = 43.5 bits (101), Expect = 9e-05
Identities = 44/137 (32%), Positives = 53/137 (38%), Gaps = 5/137 (3%)
Frame = +1
Query: 138 AAPMY---PTNSPAY--QTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPG 192
A P Y PT+ AY Q Q G QTQ P G P++ +Q P A G
Sbjct: 403 AQPSYGVPPTSQTAYGSQPQTQSGYGPPQTQKPSGTPPAYGQSQS---------PNAAGG 555
Query: 193 QTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQ 252
YG P + P + S PG P + PSG S G G +P N G+
Sbjct: 556 ----------YGQPGYPPSQPPPSGYSQPGYPPS-QPPPSGYSQPGYGQAP-QSYNSGSY 699
Query: 253 GMVRPGAPPNVRPGGAS 269
G P AP G AS
Sbjct: 700 GAGYPQAPAYSADGNAS 750
>TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {Arabidopsis
thaliana}, partial (34%)
Length = 1297
Score = 40.8 bits (94), Expect = 6e-04
Identities = 35/110 (31%), Positives = 43/110 (38%), Gaps = 1/110 (0%)
Frame = +3
Query: 129 PAPVWQNPQAA-PMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVP 187
P+PV P A PM + SP T GG A GG + G P P
Sbjct: 654 PSPVVSTPPAGGPMASSPSPVVSTPPAGGPMASSPSPAGGPPALSPAGGPSTAAGGPPAP 833
Query: 188 GAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTS 237
G G T P G A AS P ++ PG+P + + PSG S S
Sbjct: 834 GP-----GGAATSPGPGGAG---ASGPGGAAAAPGSPGSNSTAPSGNSGS 959
Score = 35.8 bits (81), Expect = 0.018
Identities = 43/147 (29%), Positives = 52/147 (35%), Gaps = 14/147 (9%)
Frame = +3
Query: 139 APMYPTNSPAYQT---QVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTG 195
+P P +SP+ P SP T P G PS P A P +
Sbjct: 486 SPRTPKSSPSPSAGGLSPPSPSPTTTTPSPSGSPPS---------------PVAIPPASS 620
Query: 196 AFPTMPSYGSAAMPTASSPLAQSSHP--GAPHN------VNLQPSGG---STSGPGSSPH 244
PT + PTASSP S P G P V+ P+GG S+ P P
Sbjct: 621 PVPT-------SGPTASSPSPVVSTPPAGGPMASSPSPVVSTPPAGGPMASSPSPAGGPP 779
Query: 245 MQQNLGAQGMVRPGAPPNVRPGGASPS 271
G G PP PGGA+ S
Sbjct: 780 ALSPAGGPS-TAAGGPPAPGPGGAATS 857
>TC88879 weakly similar to PIR|T04011|T04011 hypothetical protein T5L19.200
- Arabidopsis thaliana, partial (6%)
Length = 1573
Score = 39.7 bits (91), Expect = 0.001
Identities = 51/155 (32%), Positives = 60/155 (37%), Gaps = 13/155 (8%)
Frame = +3
Query: 135 NPQAAPMYPTNSPAYQTQVP---GGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYP 191
NPQ P YP S P G P Y GQ P A +PG +P G P
Sbjct: 693 NPQ--PTYPQASAQANYAPPQQYGKPPLYGVPPSQGQHPQSYGHPRATQPGEIPYQGPTP 866
Query: 192 GQTGAFPTMP-SYGS-----AAMPT-ASSPLAQS-SHP-GAPHNVNLQPSGGSTSG-PGS 241
Q+ P Y S AA PT S+P A SHP AP V QP G + G PG+
Sbjct: 867 AQSYGTVQQPYPYASSGPSQAAYPTYGSAPAADGYSHPQSAPGQVYAQPGGQPSYGQPGA 1046
Query: 242 SPHMQQNLGAQGMVRPGAPPNVRPGGASPSGQHYY 276
Q + + V P G+ PS Q Y
Sbjct: 1047-----QAVASYAQVGPTG------YGSYPSSQQTY 1118
>TC89983 similar to GP|14331087|emb|CAC40974. hypothetical protein {Vibrio
sp. CH-291}, partial (26%)
Length = 761
Score = 39.3 bits (90), Expect = 0.002
Identities = 29/84 (34%), Positives = 37/84 (43%)
Frame = +2
Query: 188 GAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQ 247
G Y + AF YG+ P A+SP A + PG P + G+T PG +P
Sbjct: 257 GKYTAFSQAFRIEDVYGNIGNPPATSPAAPAGTPGTP------GTPGTTGAPG-APGAPG 415
Query: 248 NLGAQGMVRPGAPPNVRPGGASPS 271
GA G GAP GASP+
Sbjct: 416 KTGAPGTT--GAPGTPNSTGASPA 481
Score = 26.9 bits (58), Expect = 8.6
Identities = 26/67 (38%), Positives = 29/67 (42%), Gaps = 2/67 (2%)
Frame = +2
Query: 154 PGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPG--AYPGQTGAFPTMPSYGSAAMPTA 211
P SPA PG P T A PG PG PG TGA T S G++ +A
Sbjct: 323 PATSPAAPAGTPG--TPGTPGTTGA--PGAPGAPGKTGAPGTTGAPGTPNSTGASPATSA 490
Query: 212 SSPLAQS 218
SS L S
Sbjct: 491 SSNLFPS 511
>TC81448 weakly similar to OMNI|NT01MC3624 toxin secretion ATP-binding
protein putative {Magnetococcus sp. MC-1}, partial (4%)
Length = 655
Score = 38.1 bits (87), Expect = 0.004
Identities = 32/91 (35%), Positives = 39/91 (42%), Gaps = 3/91 (3%)
Frame = -2
Query: 154 PGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASS 213
PGG PA VPG P A PG +P P GAFP+ P G+ A+P +
Sbjct: 336 PGGPPA--GGVPGAAFP-------ASPPGAGALPAGAPPGAGAFPSPP--GAGALPAGAP 190
Query: 214 PLAQS--SHPGAPHNV-NLQPSGGSTSGPGS 241
P A + S PGA P G PG+
Sbjct: 189 PGAGAFPSPPGAGAGAFPASPPGAGAGAPGA 97
Score = 35.0 bits (79), Expect = 0.032
Identities = 27/88 (30%), Positives = 35/88 (39%)
Frame = +2
Query: 185 PVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPH 244
P PG G+ A P G A P +P ++ PG GG+ +G +P
Sbjct: 116 PAPGGDAGKAPA-PAPGGDGKAPAP-GGAPAGKAPAPGGDGKA--PAPGGAPAGKAPAPG 283
Query: 245 MQQNLGAQGMVRPGAPPNVRPGGASPSG 272
G G PG PP P G +PSG
Sbjct: 284 -----GDAGKAAPGTPPAGGPPGVTPSG 352
Score = 31.2 bits (69), Expect = 0.46
Identities = 33/117 (28%), Positives = 45/117 (38%), Gaps = 2/117 (1%)
Frame = +2
Query: 136 PQAAPMYPTNSPAY--QTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQ 193
P AP +P + PGG+PA + PGG PG P G PG
Sbjct: 185 PGGAPAGKAPAPGGDGKAPAPGGAPAGKAPAPGGDAGK-------AAPG-TPPAGGPPGV 340
Query: 194 TGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLG 250
T P+ + AA P A++P S GA G+T+ GS ++ G
Sbjct: 341 T---PSGSASPPAATPAAAAP---KSSTGAT---------GTTAATGSGNSLKSEFG 466
>TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 -
Arabidopsis thaliana, partial (3%)
Length = 757
Score = 36.6 bits (83), Expect = 0.011
Identities = 43/156 (27%), Positives = 57/156 (35%), Gaps = 5/156 (3%)
Frame = +3
Query: 121 RDYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVR 180
+D P P V P P+ PT PG S A Q PG
Sbjct: 63 KDCPKPQPPVTVGPVPLPTPI-PTGPGGVPQPGPGASGAPQ---PG-------------- 188
Query: 181 PGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTS--- 237
PG P PG +GA P G++ P + + PG + QP G++
Sbjct: 189 PGASGAPQPGPGASGA--PQPGPGASGAPQPGPGASGAPQPGPDASGAPQPGPGASGAPQ 362
Query: 238 -GPGSSPHMQQNLGAQGMVRPG-APPNVRPGGASPS 271
GPG+S Q A G PG +P +V G+ S
Sbjct: 363 PGPGASGAPQPAPEASGAPHPGPSPTHVTTAGSGKS 470
>TC77828 similar to GP|16648867|gb|AAL24285.1 HSP associated protein like
{Arabidopsis thaliana}, partial (83%)
Length = 1587
Score = 36.6 bits (83), Expect = 0.011
Identities = 25/86 (29%), Positives = 29/86 (33%)
Frame = -3
Query: 181 PGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPG 240
PG P PG PG P P P +P PG P + P PG
Sbjct: 1162 PGMPPAPGKPPG----MPPAPGKPPGMPPAPGNPPGMPPAPGNPPGMPPAPGNPPGMPPG 995
Query: 241 SSPHMQQNLGAQGMVRPGAPPNVRPG 266
+ P M PG PP + PG
Sbjct: 994 NPPGMP----------PGNPPGMPPG 947
>TC89735 similar to GP|19569060|gb|AAL92018.1 UPF1 {Arabidopsis thaliana},
partial (18%)
Length = 1479
Score = 36.2 bits (82), Expect = 0.014
Identities = 27/93 (29%), Positives = 39/93 (41%), Gaps = 1/93 (1%)
Frame = +2
Query: 180 RPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGP 239
+P +P GA G GA P +PS GS S PH P G + + P
Sbjct: 188 QPYAIPSRGAVHGPVGAVPHVPSPGSRGFGAGRGNSGASIGNHLPHQGTQPPIGSAFNFP 367
Query: 240 G-SSPHMQQNLGAQGMVRPGAPPNVRPGGASPS 271
+P+ Q ++G + +PG N+ GA S
Sbjct: 368 ALENPNSQPSVGGP-LSQPGFANNMPVQGAGQS 463
>TC87144 similar to GP|21595622|gb|AAM66118.1 unknown {Arabidopsis
thaliana}, partial (52%)
Length = 994
Score = 35.4 bits (80), Expect = 0.024
Identities = 31/106 (29%), Positives = 41/106 (38%), Gaps = 7/106 (6%)
Frame = +3
Query: 177 HAVRPGYVPVPGAYPGQTGAFPTMPSYG----SAAMPTASSPLAQSSHPGAPHNVNLQPS 232
H + G+ PGAYP GA+P P +G P A P P + P+
Sbjct: 177 HGLGGGHGYPPGAYPPPPGAYPPPPGHGYPPHQGGYPPAGYPPPGGYPPAGYAHGGYPPA 356
Query: 233 GGSTSG-PG-SSPHMQQNLGAQGM-VRPGAPPNVRPGGASPSGQHY 275
G G PG S+PH G G G + G A+ G H+
Sbjct: 357 GYPPHGYPGPSAPHAPGPYGHAGHGAMGGGIGGLIAGAAAAYGAHH 494
>AW696480 weakly similar to SP|P03211|EBN1_ EBNA-1 nuclear protein. [strain
B95-8 Human herpesvirus 4] {Epstein-barr virus},
partial (5%)
Length = 505
Score = 34.7 bits (78), Expect = 0.041
Identities = 41/138 (29%), Positives = 49/138 (34%), Gaps = 3/138 (2%)
Frame = +1
Query: 136 PQAAP--MYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQ 193
P +AP M PTN P + G PA+ GG +P P P G
Sbjct: 28 PSSAPPGMPPTNVPP-SNLLSNGPPAFS----GGAMPG---------PPRFPGGGVQQPP 165
Query: 194 TGAFPTMPSYGSAAM-PTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQ 252
G PTM + A P +P AQS AP +QP P QQ +G
Sbjct: 166 LGP-PTMRTPAPPAQSPFNMAPPAQSPFNMAPPQGIMQPPSSPFGAPSWQTQQQQQVG-- 336
Query: 253 GMVRPGAPPNVRPGGASP 270
PP PG A P
Sbjct: 337 -------PPPTVPGSAQP 369
>TC92765 similar to EGAD|119550|127787 hypothetical protein F49E11.11
{Caenorhabditis elegans}, partial (11%)
Length = 972
Score = 33.9 bits (76), Expect = 0.070
Identities = 29/107 (27%), Positives = 44/107 (41%), Gaps = 6/107 (5%)
Frame = -2
Query: 143 PTNSPAYQTQVPGGSPAYQTQVPG------GQVPSWDLTQHAVRPGYVPVPGAYPGQTGA 196
P S +T+ P+ + + PG G V SW LT A+ P + + P +
Sbjct: 419 PCTSATARTRASFTGPSSRLRWPGPLSLSTGTVLSWSLTLTALLPASLATSSSGPSLSTE 240
Query: 197 FPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP 243
++PS T ++PLA S P + +L S GSSP
Sbjct: 239 VSSLPS-------TRTTPLASPSAFWLPPSASLSSIARS*PSSGSSP 120
>TC88753 similar to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog F9F13.90 -
Arabidopsis thaliana, partial (77%)
Length = 1273
Score = 33.9 bits (76), Expect = 0.070
Identities = 41/141 (29%), Positives = 47/141 (33%), Gaps = 8/141 (5%)
Frame = +2
Query: 129 PAPVWQNPQAAP---MYPTNSPAY----QTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRP 181
PAP PQ + YP P Q PG Q+ G P Q RP
Sbjct: 536 PAPGMMQPQISRPPVSYPGGPPVMRPPGQMPYPGHPGQGPPQMVRGPPPPMPPGQFGQRP 715
Query: 182 GYVPVPGAYPGQTGAFPTMPSYGSAAM-PTASSPLAQSSHPGAPHNVNLQPSGGSTSGPG 240
G P Q G P P YG M P + + P AP +QP PG
Sbjct: 716 G------GPPPQFGMPP--PQYGQRPMGPPPPGQMVRG--PPAPPRPGMQPP----PRPG 853
Query: 241 SSPHMQQNLGAQGMVRPGAPP 261
P + G RPG PP
Sbjct: 854 MPPPPGSGVPVYGPPRPGMPP 916
Score = 33.5 bits (75), Expect = 0.092
Identities = 44/157 (28%), Positives = 49/157 (31%), Gaps = 13/157 (8%)
Frame = +2
Query: 129 PAPVWQNPQ--AAPMYPTNSPA---YQTQV-------PGGSPAYQTQVPGGQVPSWDLTQ 176
PAPV Q A P+ PA Q Q+ PGG P + P GQ+P
Sbjct: 476 PAPVIQAQPGLAGPVRGVGGPAPGMMQPQISRPPVSYPGGPPVMR---PPGQMP------ 628
Query: 177 HAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPH-NVNLQPSGGS 235
YPG G P G P P PG P + P
Sbjct: 629 -------------YPGHPGQGPPQMVRGP---PPPMPPGQFGQRPGGPPPQFGMPPPQYG 760
Query: 236 TSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSG 272
G P Q G RPG P RPG P G
Sbjct: 761 QRPMGPPPPGQMVRGPPAPPRPGMQPPPRPGMPPPPG 871
Score = 28.9 bits (63), Expect = 2.3
Identities = 20/64 (31%), Positives = 22/64 (34%)
Frame = +2
Query: 126 PLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVP 185
P+ P Q P P P Q P G P V G P Q RPG P
Sbjct: 683 PMPPGQFGQRPGGPPPQFGMPPPQYGQRPMGPPPPGQMVRGPPAPPRPGMQPPPRPGMPP 862
Query: 186 VPGA 189
PG+
Sbjct: 863 PPGS 874
>TC78767 similar to PIR|S59117|S59117 small nuclear ribonucleoprotein U1A -
potato, partial (93%)
Length = 1455
Score = 33.1 bits (74), Expect = 0.12
Identities = 36/152 (23%), Positives = 61/152 (39%), Gaps = 21/152 (13%)
Frame = +3
Query: 35 IENMQYAVTVDVI----NTVFSAFGTVQKIAMFEK-NGQTQALIQYPDVTTAAAAREALE 89
I N+ + +D + + VFS FG + ++ F+ + QA + + DVT+A+ A ++
Sbjct: 498 INNLNEKIKIDELKKSLHAVFSQFGKILEVLAFKTLKHKGQAWVIFEDVTSASNALRQMQ 677
Query: 90 GHCIYDGGYCKLHLSYSR-HTDLNVKA---------------FSDKSRDYTVPLVPAPVW 133
G YD + + Y+R +D+ KA + K R A
Sbjct: 678 GFPFYDK---PMRIQYARTKSDVIAKAEGTFVPREKRKRHDDKAGKKRKDQNDANLAGTG 848
Query: 134 QNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVP 165
NP A Y Q PGG+ + + P
Sbjct: 849 LNPAYAGAYGATPALSQIPYPGGAKSLLPEAP 944
>TC89233 homologue to GP|18734|emb|CAA36734.1|| DNA-directed RNA polymerase
{Glycine max}, partial (21%)
Length = 648
Score = 32.7 bits (73), Expect = 0.16
Identities = 19/69 (27%), Positives = 26/69 (37%)
Frame = +3
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFP 198
+P Y SP+Y P SP+ T P S P Y P + TG P
Sbjct: 87 SPSYSPTSPSYSPTSPSYSPSSPTYSPSSPYNS------GTSPDYSPSSPQFSPSTGYSP 248
Query: 199 TMPSYGSAA 207
+ P Y ++
Sbjct: 249 SQPGYSPSS 275
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.130 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,149,884
Number of Sequences: 36976
Number of extensions: 118942
Number of successful extensions: 693
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 628
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 655
length of query: 277
length of database: 9,014,727
effective HSP length: 95
effective length of query: 182
effective length of database: 5,502,007
effective search space: 1001365274
effective search space used: 1001365274
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140025.4


