

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140024.11 + phase: 0
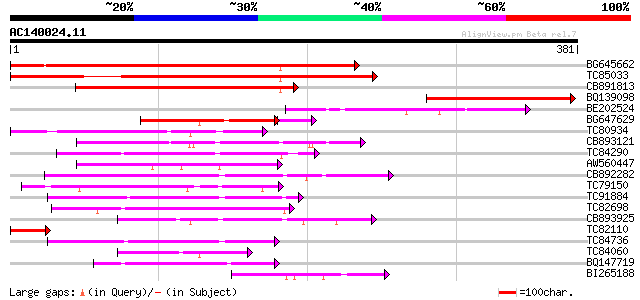
(381 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG645662 weakly similar to GP|21321208|dbj S-locus-related I {Br... 375 e-104
TC85033 weakly similar to PIR|T14419|T14419 S-locus-specific gly... 356 7e-99
CB891813 weakly similar to PIR|T14416|T144 S-locus-specific glyc... 226 1e-59
BQ139098 weakly similar to GP|22208478|gb| receptor-like kinase ... 152 3e-37
BE202524 126 1e-29
BG647629 similar to GP|9294449|dbj| receptor kinase 1 {Arabidops... 105 5e-25
TC80934 weakly similar to GP|1272349|gb|AAA97903.1| secreted gly... 81 6e-16
CB893121 weakly similar to GP|4008008|gb|A receptor-like protein... 75 3e-14
TC84290 weakly similar to GP|6554181|gb|AAF16627.1| T23J18.8 {Ar... 72 4e-13
AW560447 similar to PIR|T14520|T14 probable S-receptor kinase (E... 65 3e-11
CB892282 weakly similar to GP|20804875|dbj putative serine/threo... 62 5e-10
TC79150 similar to GP|18461304|dbj|BAB84499. putative receptor-l... 58 7e-09
TC91884 similar to GP|4741219|emb|CAB41879.1 SRK15 protein {Bras... 57 1e-08
TC82698 similar to PIR|T00534|T00534 S-receptor kinase (EC 2.7.1... 56 3e-08
CB893925 weakly similar to GP|1272349|gb|A secreted glycoprotein... 55 3e-08
TC82110 54 8e-08
TC84736 53 2e-07
TC84060 homologue to GP|1272349|gb|AAA97903.1| secreted glycopro... 53 2e-07
BQ147719 weakly similar to GP|1272349|gb|A secreted glycoprotein... 53 2e-07
BI265188 weakly similar to PIR|T05753|T057 S-receptor kinase (EC... 52 4e-07
>BG645662 weakly similar to GP|21321208|dbj S-locus-related I {Brassica
amplexicaulis}, partial (13%)
Length = 761
Score = 375 bits (963), Expect = e-104
Identities = 185/244 (75%), Positives = 207/244 (84%), Gaps = 9/244 (3%)
Frame = +1
Query: 1 MISFEIKKQVVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNN 60
MISFEIKKQVVLIYLWLWWNTT+ ICV+A +DSLKPGDKL+ S LCSKQGK+C+QF
Sbjct: 19 MISFEIKKQVVLIYLWLWWNTTS-ICVKAINDSLKPGDKLDANSNLCSKQGKYCVQFSPT 195
Query: 61 SNSDFQCLFISVNADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSP 120
++ L +SVN DYG VVW+YD NHSID +++VLSLDYSGVLKIESQ+RKPIIIYSSP
Sbjct: 196 LQNEDAHLIVSVNEDYGAVVWMYDRNHSIDLDSAVLSLDYSGVLKIESQSRKPIIIYSSP 375
Query: 121 QPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLV 180
QP NNT+AT+LD GNFVL+QF PNGS +VLWQSFDYPSDVLIP MKLGVNRKT HNWSLV
Sbjct: 376 QPINNTLATILDTGNFVLRQFHPNGSKTVLWQSFDYPSDVLIPTMKLGVNRKTAHNWSLV 555
Query: 181 ---------SDKFNLEWEPKQGELNIKKSGKVYWKSGKLKSNGLFENIPANVQSRYQYII 231
S KF+LEWEPKQGELNIKK GKVYWKSGKLKS+GLFENIPANVQ+ YQY I
Sbjct: 556 SWLTPSRPNSGKFSLEWEPKQGELNIKKRGKVYWKSGKLKSDGLFENIPANVQTMYQYTI 735
Query: 232 VSNK 235
VSN+
Sbjct: 736 VSNR 747
>TC85033 weakly similar to PIR|T14419|T14419 S-locus-specific glycoprotein -
turnip (fragment), partial (11%)
Length = 871
Score = 356 bits (914), Expect = 7e-99
Identities = 184/256 (71%), Positives = 200/256 (77%), Gaps = 9/256 (3%)
Frame = +3
Query: 1 MISFEIKKQVVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNN 60
MISFEIKKQVVLIYLWLWWNTTANICVEATSDSLKPGDK + S L SKQ
Sbjct: 99 MISFEIKKQVVLIYLWLWWNTTANICVEATSDSLKPGDKFDANSTLYSKQ---------- 248
Query: 61 SNSDFQCLFISVNADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSP 120
DYG VW+YD NHSID +++VLSLDYSGVLKIESQNRKPIIIYSSP
Sbjct: 249 --------------DYGIQVWMYDRNHSIDLDSAVLSLDYSGVLKIESQNRKPIIIYSSP 386
Query: 121 QPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLV 180
QP NNT+AT+LD GNFVLQQF PNGS +VLWQSFDYPSDVLIP MKLGVNRKTG+NWSLV
Sbjct: 387 QPINNTLATILDTGNFVLQQFHPNGSKTVLWQSFDYPSDVLIPTMKLGVNRKTGYNWSLV 566
Query: 181 ---------SDKFNLEWEPKQGELNIKKSGKVYWKSGKLKSNGLFENIPANVQSRYQYII 231
S +F+LEWEPKQGELNIKKSGKVYWKSGKLKSNGLFENIPANVQ+ Y+YII
Sbjct: 567 SWLTPSRTTSGEFSLEWEPKQGELNIKKSGKVYWKSGKLKSNGLFENIPANVQNMYRYII 746
Query: 232 VSNKDEDSFTFEVKDG 247
VSNK + ++K G
Sbjct: 747 VSNKMKIPLVLKLKIG 794
Score = 34.7 bits (78), Expect = 0.063
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 4/39 (10%)
Frame = +2
Query: 235 KDEDSFTFEVKDGKF---AQWELSSKGKLVGDDG-YIAN 269
+DEDSF+FE+KD + + W L G L D+G YI N
Sbjct: 755 QDEDSFSFEIKDRNYKNISGWTLDWAGMLTSDEGTYIGN 871
>CB891813 weakly similar to PIR|T14416|T144 S-locus-specific glycoprotein -
turnip (fragment), partial (13%)
Length = 484
Score = 226 bits (575), Expect = 1e-59
Identities = 112/160 (70%), Positives = 128/160 (80%), Gaps = 10/160 (6%)
Frame = +3
Query: 45 KLCSKQGKFCLQFGNNSNSDFQCLFISVNADYGKVVWVYDINHSIDFNTSVLSLDYSGVL 104
KLCSKQGK+C+ +S+ LFI +NADYGKVVW++D NHSID N++VLSLDYSGVL
Sbjct: 3 KLCSKQGKYCIYLKRTLDSEDAHLFIGLNADYGKVVWMHDRNHSIDLNSAVLSLDYSGVL 182
Query: 105 KIESQNRK-PIIIYSSPQPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIP 163
KIESQNRK PIIIYSSP PTNNTVATMLD GNFVLQ+ PNG+ ++LWQSFDYP+ LIP
Sbjct: 183 KIESQNRKLPIIIYSSPHPTNNTVATMLDTGNFVLQKIHPNGTKNILWQSFDYPTATLIP 362
Query: 164 MMKLGVNRKTGHNWSLV---------SDKFNLEWEPKQGE 194
MKLGVNRKTGHNWSLV S F+LEWEPK+GE
Sbjct: 363 TMKLGVNRKTGHNWSLVSWLAHSLPNSGGFSLEWEPKEGE 482
>BQ139098 weakly similar to GP|22208478|gb| receptor-like kinase {Sorghum
bicolor}, partial (5%)
Length = 674
Score = 152 bits (383), Expect = 3e-37
Identities = 66/101 (65%), Positives = 80/101 (78%), Gaps = 1/101 (0%)
Frame = +2
Query: 281 GCQKWEDIPTCREPGEMFQKKAGRPSIDNSTTYEFDVTYSYSDCKIRCWKNCSCNGFQLY 340
GCQKWEDIPTCREPGE+FQ+K GRP+I N++T E DV Y YSDCK+RCW+NC+C GF+
Sbjct: 2 GCQKWEDIPTCREPGEVFQRKTGRPNIINASTTEGDVNYGYSDCKMRCWRNCNCYGFEEL 181
Query: 341 YSNMTGCVFLSWNSTQYVDM-VPDKFYTLVKTTKSAPNSHG 380
YSN TGC++ SWNSTQ VD+ + FY LVK +K A SHG
Sbjct: 182 YSNFTGCIYYSWNSTQDVDLDDQNNFYALVKPSKPAQKSHG 304
>BE202524
Length = 564
Score = 126 bits (317), Expect = 1e-29
Identities = 73/170 (42%), Positives = 97/170 (56%), Gaps = 5/170 (2%)
Frame = +1
Query: 186 LEWEPKQGELNIKKSGKVYWKSGKLKSNGLFENIPANVQSRYQYIIVSNKDEDSFTFEVK 245
L EP EL IK KVYW+SGKLK+N FE I + +VSN +E+ F++ +
Sbjct: 70 LNGEPNIKELVIKHREKVYWRSGKLKNN-TFEYISGK---DIKVNVVSN*NEEYFSYTTQ 237
Query: 246 -DGKFAQWELSSKGKLVGDDG--YIANADMCYGYNSDGGCQKWED--IPTCREPGEMFQK 300
+ W L G+L+ + +I ADMCYGYN++ GCQKW D IPTCR PG+ F
Sbjct: 238 NEDSLTVWTLLETGQLIDREASDHIGRADMCYGYNTNDGCQKWGDAEIPTCRNPGDKFDS 417
Query: 301 KAGRPSIDNSTTYEFDVTYSYSDCKIRCWKNCSCNGFQLYYSNMTGCVFL 350
K P+ + + + +Y SDC+ CW+NCSC GF YSN TG V L
Sbjct: 418 KIVYPN-EKIEYHILNSSYGISDCQDMCWRNCSCFGFGNLYSNGTGWVIL 564
>BG647629 similar to GP|9294449|dbj| receptor kinase 1 {Arabidopsis
thaliana}, partial (5%)
Length = 539
Score = 105 bits (261), Expect(2) = 5e-25
Identities = 55/95 (57%), Positives = 72/95 (74%), Gaps = 2/95 (2%)
Frame = +1
Query: 89 IDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNT--VATMLDAGNFVLQQFLPNGS 146
+ +++VLSL+ SG LKIES+ KPI +Y+S QP N + VAT+LD+GNFVL+ N
Sbjct: 79 LTMDSAVLSLNQSGALKIESKFGKPITLYASHQPLNRSTNVATLLDSGNFVLKDTQKN-- 252
Query: 147 MSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVS 181
+VLWQSFD+P+D L+P MKLG+N KTG NWSLVS
Sbjct: 253 -TVLWQSFDHPTDSLLPGMKLGLNHKTGENWSLVS 354
Score = 26.9 bits (58), Expect(2) = 5e-25
Identities = 14/26 (53%), Positives = 15/26 (56%)
Frame = +2
Query: 181 SDKFNLEWEPKQGELNIKKSGKVYWK 206
S F L+WEP EL IK KVY K
Sbjct: 386 SGPFRLDWEPIGKELVIKHREKVY*K 463
>TC80934 weakly similar to GP|1272349|gb|AAA97903.1| secreted glycoprotein 3
{Ipomoea trifida}, partial (24%)
Length = 732
Score = 81.3 bits (199), Expect = 6e-16
Identities = 63/178 (35%), Positives = 93/178 (51%), Gaps = 5/178 (2%)
Frame = +2
Query: 1 MISFEIKKQVVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNN 60
M+ KK ++L +L TT ++ D+L +++ L S +G F + F +
Sbjct: 95 MVDISFKKLIILFFLSSSLRTTTSL------DTLSLNQSIHHGQSLVSAKGTFEVGFFSP 256
Query: 61 SNSDFQCLFISV-NADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSS 119
NS + L + N VVWV + + N VL L+ +GVL+I N K I+SS
Sbjct: 257 GNSRGRYLGMWYKNLTPLTVVWVANRETPLHNNLGVLKLNENGVLEI--LNGKNYAIWSS 430
Query: 120 P---QPTNNTV-ATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKT 173
+P NN++ A +LD GN VL+ N + ++LWQSFDYPSD +P MKLG N T
Sbjct: 431 NASNRPANNSISAQLLDNGNLVLK----NSANNILWQSFDYPSDTFLPGMKLGWNLVT 592
>CB893121 weakly similar to GP|4008008|gb|A receptor-like protein kinase
{Arabidopsis thaliana}, partial (11%)
Length = 724
Score = 75.5 bits (184), Expect = 3e-14
Identities = 65/215 (30%), Positives = 104/215 (48%), Gaps = 21/215 (9%)
Frame = +1
Query: 46 LCSKQGKFCLQFGNNSNSDFQCLFI-SVNADYGKVVWVYDINHSIDFNTSVLSLDYSGVL 104
L SK F L F S + + I N VVWV + + I+ + +LS+D +G L
Sbjct: 13 LVSKSKTFALGFFTPGKSASRYVGIWYYNLPIQTVVWVANRDAPINDTSGILSIDPNGNL 192
Query: 105 KIESQNRKPIIIYSS----PQP----TNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDY 156
I N I I+S+ PQ TN +A + D N VL + N + +V+W+SFD+
Sbjct: 193 VIH-HNHSTIPIWSTDVSFPQSQRNSTNAVIAKLSDIANLVL---MINNTKTVIWESFDH 360
Query: 157 PSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNIKKS--GK----------VY 204
P+D L+P +K+G NRKT +W L S + + +P +G ++ S GK +
Sbjct: 361 PTDTLLPYLKIGFNRKTNQSWFLQS--WKTDDDPGKGAFTVEFSTIGKPQLFMYNHNLPW 534
Query: 205 WKSGKLKSNGLFENIPANVQSRYQYIIVSNKDEDS 239
W++G LF +P + + + +DE+S
Sbjct: 535 WRAGHWNGE-LFAGVPNMKRDMETFNVSFVEDENS 636
>TC84290 weakly similar to GP|6554181|gb|AAF16627.1| T23J18.8 {Arabidopsis
thaliana}, partial (6%)
Length = 873
Score = 72.0 bits (175), Expect = 4e-13
Identities = 57/195 (29%), Positives = 90/195 (45%), Gaps = 18/195 (9%)
Frame = +1
Query: 32 DSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNADYGKVVWVYDINHSIDF 91
D++ + L SK G F L F + NS + + I + ++WV + N S++
Sbjct: 157 DTITSSQFIKDPETLLSKDGNFTLGFFSPKNSTNRYVGIWWKSQ-STIIWVANRNRSLND 333
Query: 92 NTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTV-ATMLDAGNFVLQQFLPNGSMSVL 150
+ ++++ G L + + ++ I + T+NT+ A D GN VL L + + S+L
Sbjct: 334 SNGIITISEDGNLAVLNGQKQVIWSSNLSNTTSNTIEAQFSDYGNLVL---LESTTGSIL 504
Query: 151 WQSFDYPSDVLIPMMKLGVNRKTGHNWSLVS-----------------DKFNLEWEPKQG 193
WQSF PSD L+P MKL N++TG L S D+ NL
Sbjct: 505 WQSFQKPSDTLLPGMKLTSNKRTGEKVQLTSWKNPSDPSVGSFSISFIDRINLH------ 666
Query: 194 ELNIKKSGKVYWKSG 208
EL I + YW+SG
Sbjct: 667 ELFIFNETQPYWRSG 711
>AW560447 similar to PIR|T14520|T14 probable S-receptor kinase (EC 2.7.1.-)
SFR3 precursor - wild cabbage, partial (12%)
Length = 641
Score = 65.5 bits (158), Expect = 3e-11
Identities = 53/149 (35%), Positives = 76/149 (50%), Gaps = 11/149 (7%)
Frame = +1
Query: 46 LCSKQGKFCLQFGNNSNSDFQCLFISV-NADYGKVVWVYDINHSIDFNTS---VLSLDYS 101
L S G F L F +S + + I N ++VWV + ++ I NTS +L +
Sbjct: 76 LVSNDGTFELGFFTPGSSTNRYVGIWYKNIPKRRIVWVANRDNPIKDNTSNSTMLIMSND 255
Query: 102 GVLKIESQNRKPI-----IIYSSPQPTNNTVATMLDAGNFVLQ--QFLPNGSMSVLWQSF 154
G L+I + N + + I S T++ VA +LD GNFV++ S + LWQ F
Sbjct: 256 GNLEILTNNNQTLVWSTNITTQSLSTTSSHVAQLLDNGNFVIKANNNTDQQSNNFLWQGF 435
Query: 155 DYPSDVLIPMMKLGVNRKTGHNWSLVSDK 183
D+P D L+P MKLG + KTG N L S K
Sbjct: 436 DFPCDTLLPDMKLGWDLKTGLNRQLTSWK 522
>CB892282 weakly similar to GP|20804875|dbj putative serine/threonine kinase
{Oryza sativa (japonica cultivar-group)}, partial (6%)
Length = 820
Score = 61.6 bits (148), Expect = 5e-10
Identities = 67/251 (26%), Positives = 105/251 (41%), Gaps = 16/251 (6%)
Frame = +2
Query: 24 NICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISV-NADYGKVVWV 82
N + T ++ P + Y L S G + F N +S Q I N +VWV
Sbjct: 20 NFSTQKTFTTIAPNQFMQYGDTLVSAAGMYEAGFFNFGDSQRQYFGIWYKNISPRTIVWV 199
Query: 83 YDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFL 142
+ N +T++L L+ G L I ++ I + + +V + D+GN VL+
Sbjct: 200 ANRNTPTQNSTAMLKLNDQGSLVIVDGSKGIIWSSNISRIVVKSVVQLFDSGNLVLKDA- 376
Query: 143 PNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNIK---- 198
S + LW+SFDYP + + MKL N TG L S K +P +GE + K
Sbjct: 377 --NSQNFLWESFDYPGNTFLAGMKLKSNLVTGPYRYLTSWKD--PQDPAEGECSYKIDTH 544
Query: 199 --------KSGKVYWKSGKLKSNG-LFENIP-ANVQSRYQYIIVSNKDEDSFTFEVKDGK 248
K KV ++ G NG LF + ++ + +V E S+ +E +
Sbjct: 545 GFPQLVTAKGAKVLYRGGSW--NGFLFTGVSWQRLRRVLNFSVVVTDKEFSYQYETFNSF 718
Query: 249 F-AQWELSSKG 258
+W L S G
Sbjct: 719 INTRWVLDSYG 751
>TC79150 similar to GP|18461304|dbj|BAB84499. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(7%)
Length = 1077
Score = 57.8 bits (138), Expect = 7e-09
Identities = 53/188 (28%), Positives = 89/188 (47%), Gaps = 12/188 (6%)
Frame = +3
Query: 9 QVVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSK---LCSKQGKFCLQFGNNSNSDF 65
QV + L+ TT++ ++T++ L G L+ + L S G F F ++ F
Sbjct: 63 QVFFLLFLLFTKTTSS---QSTTNILPQGSSLSVEKSNNTLISSNGDFSAGFLPVGDNAF 233
Query: 66 QCLFISVNADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYS----SPQ 121
+ +VW+ + + ++ S LSL +G L + +RK I+S SP
Sbjct: 234 CFAVYFTKSKQPTIVWMANRDQPVNGKHSKLSLFKNGNLILTDADRKRTPIWSTSSFSPF 413
Query: 122 PTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLG-----VNRKTGHN 176
P + + GN VL NG++S+LWQSFD+P+D L+P ++ V+ K+ N
Sbjct: 414 PLQ---LKLQNNGNLVLST--TNGNISILWQSFDFPTDTLLPGQEINERATLVSSKSETN 578
Query: 177 WSLVSDKF 184
+S KF
Sbjct: 579 YSSGFYKF 602
>TC91884 similar to GP|4741219|emb|CAB41879.1 SRK15 protein {Brassica
oleracea}, partial (3%)
Length = 1156
Score = 57.0 bits (136), Expect = 1e-08
Identities = 42/172 (24%), Positives = 79/172 (45%)
Frame = +3
Query: 26 CVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNADYGKVVWVYDI 85
C A +D++ L + S L F + NS + L I + + W+ +
Sbjct: 612 CYSAVNDTITSSKLLKDNETITSNNTDLKLGFFSPLNSPNRYLGIWYINETNNI-WIANR 788
Query: 86 NHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLPNG 145
+ + + ++++ +G L I ++ II ++ + N+ A + DAGN +L+
Sbjct: 789 DQPLKDSNGIVTIHKNGNLVILNKPNGSIIWSTNISSSTNSTAKLDDAGNLILRDI---N 959
Query: 146 SMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNI 197
S + +W SF +PSD +P MK+ N+ TG + V+ K + +P G I
Sbjct: 960 SGATIWDSFTHPSDSAVPSMKIASNKVTGKQIAFVARK--SDNDPSSGHFTI 1109
>TC82698 similar to PIR|T00534|T00534 S-receptor kinase (EC 2.7.1.-)
T20K24.15 precursor - Arabidopsis thaliana, partial
(14%)
Length = 675
Score = 55.8 bits (133), Expect = 3e-08
Identities = 47/176 (26%), Positives = 84/176 (47%), Gaps = 13/176 (7%)
Frame = +3
Query: 29 ATSDSLKPGDKLNYKSKLCSKQGKFCLQF---GNNSNSDFQCLFISVNADYGKVVWVYDI 85
A + ++ L+ L S+ G F L F GN+SN + V +VWV +
Sbjct: 135 ALTTTISAKQSLSGDQTLISEGGIFELGFFKPGNSSNYYIGIWYKKVIQQ--TIVWVANR 308
Query: 86 NHSI-DFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLPN 144
++ + D NT+ L + ++ + +++ + +++ VA +LD GN VL+ +
Sbjct: 309 DNPVSDKNTATLKISDGNLVILNESSKQVWSTNMNVPKSDSVVAMLLDTGNLVLKNRPND 488
Query: 145 GSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDK---------FNLEWEPK 191
+ LWQSFD+P+D +P K+ ++ KT L S K F+LE +P+
Sbjct: 489 DVLDSLWQSFDHPADTWLPGGKIKLDNKTKKPQYLTSWKNRKDPATGLFSLELDPE 656
>CB893925 weakly similar to GP|1272349|gb|A secreted glycoprotein 3 {Ipomoea
trifida}, partial (19%)
Length = 715
Score = 55.5 bits (132), Expect = 3e-08
Identities = 52/190 (27%), Positives = 85/190 (44%), Gaps = 16/190 (8%)
Frame = +2
Query: 73 NADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSP---QPTNNTVAT 129
N +VVWV + ++ I +S L + L + +N+ +++S+ + ++ +A
Sbjct: 20 NIPVRRVVWVANRDNPIKDTSSKLIISQDRNLVLLDKNQS--LLWSTNATIEKVSSPIAQ 193
Query: 130 MLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEW- 188
+LD GN VL+ G LWQSFDYP D ++ MK G +++ N SLV+ K W
Sbjct: 194 LLDDGNLVLKN---GGEEHFLWQSFDYPCDTILSGMKAGWDKRKDLNRSLVAWK---NWD 355
Query: 189 EPKQGELN-----IKKSGKVYWKS-GKLKSNGLFEN------IPANVQSRYQYIIVSNKD 236
+P + WK KL G + I Y Y V+N+D
Sbjct: 356 DPSSSDFTSAMVLTPNPESFIWKGLTKLYRTGPWTGPRSSGVIGLTENPLYDYEFVNNQD 535
Query: 237 EDSFTFEVKD 246
E + F +K+
Sbjct: 536 EVYYLFTLKN 565
>TC82110
Length = 685
Score = 54.3 bits (129), Expect = 8e-08
Identities = 23/27 (85%), Positives = 25/27 (92%)
Frame = +1
Query: 1 MISFEIKKQVVLIYLWLWWNTTANICV 27
MISF+IKKQVVLIYLWLWW T+ NICV
Sbjct: 55 MISFKIKKQVVLIYLWLWWITSTNICV 135
>TC84736
Length = 599
Score = 53.1 bits (126), Expect = 2e-07
Identities = 37/157 (23%), Positives = 72/157 (45%), Gaps = 1/157 (0%)
Frame = +1
Query: 26 CVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFI-SVNADYGKVVWVYD 84
C A +D++ L + S F L + NS + L I +N +W+ +
Sbjct: 142 CNSAINDTITSSKSLKDNETITSNNTNFKLGLFSPLNSTNRYLGIWYINKTNN--IWIAN 315
Query: 85 INHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLPN 144
+ + + ++++ G I ++ II ++ + N+ A + D+GN +L+
Sbjct: 316 RDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNSTAQLADSGNLILRDI--- 486
Query: 145 GSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVS 181
S + +W SF +P+D +P M++ N+ TG S VS
Sbjct: 487 SSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVS 597
>TC84060 homologue to GP|1272349|gb|AAA97903.1| secreted glycoprotein 3
{Ipomoea trifida}, partial (8%)
Length = 673
Score = 53.1 bits (126), Expect = 2e-07
Identities = 37/99 (37%), Positives = 55/99 (55%), Gaps = 8/99 (8%)
Frame = +2
Query: 73 NADYGK-VVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNT----- 126
N D+ K VVWV + N + T+ L L +G L I +++ K I +SS Q N+
Sbjct: 197 NIDHDKTVVWVANRNTPLQNPTAFLKLTNTGNLIIINESNKTI--WSSNQTNQNSTLNTN 370
Query: 127 -VATMLDAGNFVLQ-QFLPNGSMSVLWQSFDYPSDVLIP 163
+ +LD+GN V+ + N + LWQSFDYP+D L+P
Sbjct: 371 PILQLLDSGNLVVTTEPNENDPTNFLWQSFDYPTDTLLP 487
>BQ147719 weakly similar to GP|1272349|gb|A secreted glycoprotein 3 {Ipomoea
trifida}, partial (15%)
Length = 693
Score = 53.1 bits (126), Expect = 2e-07
Identities = 34/125 (27%), Positives = 61/125 (48%)
Frame = +3
Query: 57 FGNNSNSDFQCLFISVNADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIII 116
FGN++N F + +++ +VW+ + + + ++ VL+L G L I I
Sbjct: 105 FGNSNNQYFGVWYKNISPK--TLVWIANRDVPLGNSSGVLNLTDKGTLVIVDSKEVTIWS 278
Query: 117 YSSPQPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHN 176
++ T+ +L++GN +++ + +LWQSFD P D L+P M + N G
Sbjct: 279 SNTSTTTSKPSLQLLESGNLIVKDEIDPDK--ILWQSFDLPGDTLLPGMSIRTNLVNGDY 452
Query: 177 WSLVS 181
LVS
Sbjct: 453 KGLVS 467
>BI265188 weakly similar to PIR|T05753|T057 S-receptor kinase (EC 2.7.1.-)
M4I22.100 precursor - Arabidopsis thaliana, partial (6%)
Length = 559
Score = 52.0 bits (123), Expect = 4e-07
Identities = 42/120 (35%), Positives = 59/120 (49%), Gaps = 14/120 (11%)
Frame = +1
Query: 150 LWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKF-------NLEWEP---KQGELNIKK 199
LWQSFDYPSD ++P MKLG N +T + S K +L W E + +
Sbjct: 109 LWQSFDYPSDTILPGMKLGWNLRTHTERRMTSWKSPDDPSPGDLYWGSLLYNYPEQYLMQ 288
Query: 200 SGKVYWKSGK---LKSNGLFENIPANVQSRYQYIIVSNKDEDSFTFE-VKDGKFAQWELS 255
K Y + G L +G+ + P NV Y Y VSNKDE +T+ + D ++ EL+
Sbjct: 289 GTKKYVRVGPWNGLHFSGVPDQKPNNV---YAYNFVSNKDEIYYTYSMLNDSVISRMELN 459
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,383,664
Number of Sequences: 36976
Number of extensions: 233727
Number of successful extensions: 1471
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 1428
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1453
length of query: 381
length of database: 9,014,727
effective HSP length: 98
effective length of query: 283
effective length of database: 5,391,079
effective search space: 1525675357
effective search space used: 1525675357
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140024.11


